IMP is a library for solving a a wide variety of molecular structures and dynamics using many different data sources. As a result, it provides a great deal of flexibility. In order to best make the required decisions about how to use IMP to solve a particular problem, it is useful to understand the overall structure of IMP.
IMP proceeds in a five stage iterative process
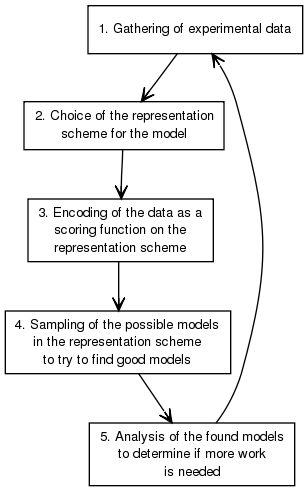
IMP provides a large number of functionality to facilitate this process. Links to representative classes are given for future reference.
IMP is via a collection of entities called particles (IMP::Particle objects). Each particle can contain one or more of the following sets of data Cartesian coordinates (IMP::core::XYZ)
Cartesian coordinates (IMP::core::XYZ) sphere (IMP::core::XYZR)
sphere (IMP::core::XYZR) atom information such as type, element, mass (IMP::atom::Atom)
atom information such as type, element, mass (IMP::atom::Atom) residue type information (IMP::atom::Residue)
residue type information (IMP::atom::Residue) chain IDs (IMP::atom::Chain)
chain IDs (IMP::atom::Chain) domain extents (IMP::atom::Domain)
domain extents (IMP::atom::Domain) mass, in daltons (IMP::atom::Mass)
mass, in daltons (IMP::atom::Mass) charge (IMP::atom::Charged)
charge (IMP::atom::Charged) relationships between parts of molecules (IMP::atom::Hierarchy)
relationships between parts of molecules (IMP::atom::Hierarchy)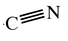 bonds (IMP::atom::Bond and IMP::atom::Bonded)
bonds (IMP::atom::Bond and IMP::atom::Bonded)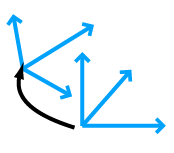 rigid body coordinate frames (IMP::core::RigidBody)
rigid body coordinate frames (IMP::core::RigidBody)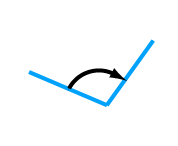 bond angle (IMP::atom::Angle)
bond angle (IMP::atom::Angle)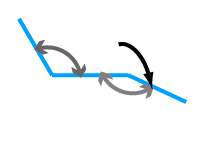 bond angle (IMP::atom::TorsionAngle)
bond angle (IMP::atom::TorsionAngle)IMP can enforce relationships between particles: all of the members of a rigid body move along with the rigid body (IMP::core::RigidBody, IMP::core::RigidMember)
all of the members of a rigid body move along with the rigid body (IMP::core::RigidBody, IMP::core::RigidMember) a centroid particle has Cartesian coordinates computed from the centroid of another set (IMP::core::Centroid)
a centroid particle has Cartesian coordinates computed from the centroid of another set (IMP::core::Centroid) a cover particle has a sphere containing another set of particles (IMP::core::Cover)
a cover particle has a sphere containing another set of particles (IMP::core::Cover)IMP the scoring function is the sum terms, each of which is computed by an IMP::Restraint object. The scoring function terms can be based on things like how close a distance is to the measured value (IMP::core::DistanceRestraint, IMP::core::DistancePairScore, IMP::core::RigidBodyDistancePairScore, IMP::core::SphereDistancePairScore)
how close a distance is to the measured value (IMP::core::DistanceRestraint, IMP::core::DistancePairScore, IMP::core::RigidBodyDistancePairScore, IMP::core::SphereDistancePairScore) how well the model fits a density map (IMP::em::FitRestraint)
how well the model fits a density map (IMP::em::FitRestraint) how close the volume of a molecule is to the expected value (IMP::core::VolumeRestraint)
how close the volume of a molecule is to the expected value (IMP::core::VolumeRestraint) excluded volume (steric clash) (IMP::core::ExcludedVolumeRestraint)
excluded volume (steric clash) (IMP::core::ExcludedVolumeRestraint) connectivity of a subcomplex (IMP::core::ConnectivityRestraint)
connectivity of a subcomplex (IMP::core::ConnectivityRestraint) the fit of the SAXS cure of a complex to a measured one (IMP::saxs::Restraint)
the fit of the SAXS cure of a complex to a measured one (IMP::saxs::Restraint) statistical potentials (IMP::atom::ProteinLigandRestraint)
statistical potentials (IMP::atom::ProteinLigandRestraint) torsion angles or bond angles (IMP::core::TorsionAngleRestraint, IMP::core::AngleRestraint)
torsion angles or bond angles (IMP::core::TorsionAngleRestraint, IMP::core::AngleRestraint) symmetry: (IMP::core::TransformedDistancePairScore)
symmetry: (IMP::core::TransformedDistancePairScore)IMP provides two sampling protocols, IMP::core::MCCGSampler which uses a combination of IMP::core::MonteCarlo and IMP::core::ConjugateGradients with randomized starting conformations and IMP::domino::DominoOptimizer which uses a graph based inference algorithm. Sampling is an iterative process that tends to be structured as follows:
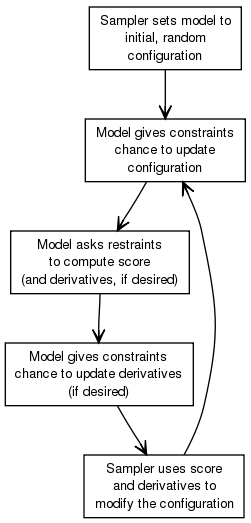
IMP provides a variety of tools to help display the conformations, in IMP::display, and to cluster them, in IMP::statistics. Display capabilities includeKnowledge about the system being modeled enters the process at all stages, but a few need extra note:
Coming up with the right choices for representation, scoring and sampling for a given system typically takes a few iterations and trial and error. IMP provides tools to help monitor how things are performing.
IMP can produce logged information to help understand what is going on. The amount of logging information produced can be controlled globally using the IMP.set_log_level() function and passing it one of the IMP::LogLevel values. In addition, restraints, samplers, constraints (and all objects which inherit from IMP::Object) have an internal log level that overrides the global one. To set that call the IMP::Object::set_log_level() function on that object. Setting the log level to IMP::VERBOSE will produce a huge amount of information during a typical sampling run. IMP::TERSE is generally better. Make sure to set it to at least IMP::WARNING to make sure that you don't miss any important warnings.
IMP can perform a lot of checks that it is being used correctly as well as that it is behaving correctly. The checks being performed are controlled by the IMP::set_check_level() function call. Set the check level to IMP::USAGE to make sure that all parameters passed are correct. Set it to IMP::USAGE_AND_INTERNAL if you are developing new restraints or sampling protocols or are worried that IMP is malfunctioning.
IMP supports I/O to and from a variety of formats. To preserve the maximum amount of information, one can use the IMP::read_model() and IMP::write_model() methods to save and load a whole model to a human (and machine) readable file. See also the IMP::display module for geometry output and IMP::atom for biological formats.IMP is organized around a number of core concepts. Representation is handled via a collection of IMP::Particle objects. Each has a set of arbitrary attributes (such as an x coordinate or a mass). In order to make particles more friendly, we provide decorators which, guess what, decorate, an existing particle to provide a higher level interface to manipulate the attributes of a particle. See the IMP::Decorator page for more details.
IMP provides containers in order to aid managing sets of particles. These inherit from IMP::Container (notice that IMP::Particle objects are containers and can contain lists of particles). A container could be as simple as an IMP::container::ListSingletonContainer which simply stores a list of particles. However, it could also be more involved, such as the IMP::container::ClosePairContainer which keeps track of all pairs of particles which close to one another in space. It can be used to implemented non-bonded operations for example.
Scoring is handled by a collection of IMP::Restraint objects. Each of these keeps a list of particles and scores those particles based on how well they fit some sort of data.
The IMP::Model manages the set of all particles in the representation along with the set of all restraints scoring them and constraints acting on them. It provides one central function, IMP::Model::evaluate() which computes the score of the current conformation.
One final representation concept is that of a constraint. These are implemented as IMP::Constraint objects. They maintain some hard invariant of the representation. Examples include, keeping a rigid body rigid, or ensuring that the IMP::container::ClosePairContainer really contains all close pairs. Constraints are updated as part of the IMP::Model::evaluate(). This means that the constraint does not necessarily hold except during score evaluation. In order to ensure that all constraints hold, call IMP::Model::evaluate() before inspecting the particles.
On the sampling side, there are two main concepts, that of an optimizer and that of a sampler. An optimizer (IMP::Optimizer), takes the current conformation of the IMP::Model and modifies it (typically in an attempt to make it score better). A sampler uses variety of optimizers and other methods to perform a non-local search for good scoring conformations, which are then stored as part of an IMP::ConfigurationSet.
IMP. They are all are in Python, but the C++ code is nearly the same.Each module has an examples page linked from its main page.
import IMP.core
def create_model_and_particles():
m= IMP.Model()
sc= IMP.container.ListSingletonContainer()
b= IMP.algebra.BoundingBox3D(IMP.algebra.Vector3D(0,0,0),
IMP.algebra.Vector3D(10,10,10))
for i in range(0,100):
p= IMP.Particle(m)
sc.add_particle(m)
d=IMP.core.XYZR.setup_particle(p, IMP.algebra.Sphere3D(IMP.algebra.get_random_vector_in(b), 1))
d.set_coordinates_are_optimized(True)
return (m, sc)
import IMP.example (m,c)=IMP.example.create_model_and_particles() uf= IMP.core.Harmonic(0,1) df= IMP.core.DistancePairScore(uf) r= IMP.core.PairRestraint(df, IMP.ParticlePair(c.get_particle(0), c.get_particle(1))) m.add_restraint(r)
import IMP.example (m,c)=IMP.example.create_model_and_particles() # this container lists all pairs that are close at the time of evaluation nbl= IMP.container.ClosePairContainer(c, 0,2) h= IMP.core.HarmonicLowerBound(0,1) sd= IMP.core.SphereDistancePairScore(h) # use the lower bound on the inter-sphere distance to push the spheres apart nbr= IMP.container.PairsRestraint(sd, nbl) m.add_restraint(nbr) # alternatively, one could just do r = IMP.core.ExcludedVolumeRestraint(c) m.add_restraint(r) # get the current score print m.evaluate(False)
import IMP.atom
m= IMP.Model()
prot= IMP.atom.read_pdb(IMP.atom.get_example_path("example_protein.pdb"), m)
bds= IMP.atom.get_internal_bonds(prot)
bl= IMP.container.ListSingletonContainer(bds.get_particles())
h= IMP.core.Harmonic(0,1)
bs= IMP.atom.BondSingletonScore(h)
br= IMP.container.SingletonsRestraint(bs, bl)
m.add_restraint(br)
print m.evaluate(False)
import IMP.example
import IMP.statistics
(m,c)=IMP.example.create_model_and_particles()
ps= IMP.core.DistancePairScore(IMP.core.HarmonicLowerBound(1,1))
r= IMP.container.PairsRestraint(ps, IMP.container.ClosePairContainer(c, 2.0))
m.add_restraint(r)
# we don't want to see lots of log messages about restraint evaluation
m.set_log_level(IMP.WARNING)
# the container (c) stores a list of particles, which are alse XYZR particles
# we can construct a list of all the decorated particles
xyzrs= IMP.core.XYZRsTemp(c.get_particles())
s= IMP.core.MCCGSampler(m)
s.set_number_of_attempts(10)
# but we do want something to watch
s.set_log_level(IMP.TERSE)
# find some configurations which move the particles far apart
configs= s.get_sample();
for i in range(0, configs.get_number_of_configurations()):
configs.set_configuration(i)
# print out the sphere containing the point set
# - Why? - Why not?
sphere= IMP.core.get_enclosing_sphere(xyzrs)
print sphere
# cluster the solutions based on their coordinates
e= IMP.statistics.ConfigurationSetXYZEmbedding(configs, c)
# of course, this doesn't return anything of interest since the points are
# randomly distributed, but, again, why not?
clustering = IMP.statistics.get_lloyds_kmeans(e, 3, 1000)
for i in range(0,clustering.get_number_of_clusters()):
# load the configuration for a central point
configs.set_configuration(clustering.get_cluster_representative(i))
sphere= IMP.core.get_enclosing_sphere(xyzrs)
print sphere
IMP is grouped into modules, each with its own namespace (in C++) or package (in Python). For example, the functionality for IMP::core can be found like in C++ and IMP.core.XYZ(p)
A module contains classes, methods and data which are related and controlled by a set of authors. The names of the authors, the license for the module, its version and an overview of the module can be found on the module main page (eg IMP::example). See the "Modules" tab above for a complete list of modules in this version of IMP.
Modules are either grouped based on types of experimental data (eg IMP::em) or based on shared functionality (IMP::core or IMP::container).
IMP can be used from both C++ and Python. We recommend that you:IMP classes together to handle your data and resulting structuresIMP classes in C++If you are new to programming you should check out a general python introduction such as the official introduction to Python and Python 101. Users who have programmed but are not familiar with Python should take a look at Dive into Python, especially chapters 1-6, and 15-18.
While effort has been made to ensure that the interfaces are the same between the two languages, a number of differences remain due to differences in the languages and limitations of the program used to generate the connection between the two languages. Key differences are
foos_begin(), foos_end(), we provide a method get_foos() which can be used with python foreach loops.Particle* with a Decorator and the decorator will automatically be converted. It will not on the python side. Similarly for converting between ParticlesTemp and Particles.IMP exceptions are exposed as identical Python exception classes. The class hierarchy is similar (e.g. all exceptions derive from IMP::Exception, so "except IMP.Exception" will catch all IMP exceptions), except for convenience some generic IMP exceptions also derive from their standard Python equivalents (e.g. IMP.IndexException derives from the standard Python IndexError as well as IMP::Exception). Thus, an IMP::IndexException could be caught in Python most specifically with "except IMP.IndexException" but also with "except IMP.Exception" or "except IndexError".IMP also uses reference counting on the C++ side so that memory managment works naturally across the language barrier. See IMP::RefCounted for a detailed description of how to do IMP reference counting in C++.IMP.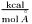 for forces/derivatives
for forces/derivatives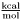 for energies
for energiesAnything that breaks from these conventions must be labeled clearly and accompanied by an explaination of why the normal units could not be used.
Name are passed using the type Names. For example, a bunch of IMP::algebra::Vector3D objects are passed using a IMP::algebra::Vector3Ds type, and a bunch of IMP::Restraint objects is passed using IMP::Restraints (or, equivalently IMP::RestraintsTemp).
IMP exceptions to report errors. See IMP::Exception for a list of existing exceptions. These C++ exceptions are mapped onto the normal python exception types.IMP classes are divided into two typesconst&). Examples include IMP::algebra::VectorD, collections such as IMP::Restraints or IMP::RestraintsTemp, or decorators, such as IMP::core::XYZ. In fact, in IMP, anything that does not inherit from IMP::RefCounted is a value class.
IMP, these classes all inherit from IMP::Object. For example always do things like this in C++: IMP::Pointer<IMP::Model> m= new IMP::Model(); IMP_NEW(Model, m, ()); // a macro which expands to the above
NamesTemp instead of a Names.Python does not have this distinction.
A few classes in IMP are designed for fast, low level use. Their default constructor leaves them in an unspecified state. This is similar to the built in types in C++ (int, double). For example
IMP::algebra::VectorD<3> v; // the vector has unknown coordinates std::cout << v << std::endl; // illegal v= IMP::algebra::VectorD<3>(0,1,2); // now we can use v
Unless the documentation says otherwise, all value class object in IMP can be compared with other equivalent objects based on their contents. Object class objects allow checking of equality to see if they are the same object (not whether two have the same state). In C++, this is done by comparing the pointers.
const method show(std::ostream&), which writes some basic information about the object to the supplied stream. In addition, on the C++ side, all objects support standard output to stream via <<. In addition, all objects support __str__ in python so that they can be printed and displayed.CamelCase, for example class SpecialVector' IMP, Name, there is a type Names which is used to pass a list of objects of type Name. Names look like an std::vector in C++ or a list in Python. Sometimes, for efficiency, a NamesTemp is passed instead (see when to use Temp values for the reason). Names will be converted into NamesTemp without cost, so the distinction should not matter for the caller.separated_by_underscores, for example void SpecialVector::add_constant(int the_constant)' set_ value object or which return an existing object class object begin with get_. No arguments of such functions are modified.class object start with create_.#define) begin with IMP_ IMP can be performed in two different ways....Incremental scoring works as follows:
To set it up call IMP::Model::set_is_incremental() with the value true. This
When evaluate is called during optimization
A IMP::Restraint is an incremental restraint if IMP::Restraint::get_is_incremental() returns true. For such restraints, IMP::Restraint::incremental_evaluate() is called instead of IMP::Restraint::evaluate().
Whenever a particle is changed is marked as dirty, so that IMP::Particle::get_is_changed() returns true.
A (perhaps partial) list of classes which benefit from incremental evaluation is:
IMP, please report it on the IMP bug tracker. This will ensure it does not get lost. The best way to report a bug is to provide a short script file that demonstrates the problem.IMP can be found in the installation instructions.There are a few areas of core functionality that have already been mentioned.
Then look through the examples which can be found linked from the page of each module.
There are a variety of useful base classes which are used to provide most functionality. They are:
There are a few blocks of functionality that cut across modules. They include
When programming with IMP, one of the more useful pages is the modules list.
For general help, you can use the imp-users mailing list.