IMP represents molecular structures using the Hierachy decorator. Using it, molecules and collections of molecules are represented using a hierarchy (or tree) where the level of detail increases as you move further from the root. The leaves of the tree have coordinates, as can internal nodes if the molecules are represented at multiple resolutions.
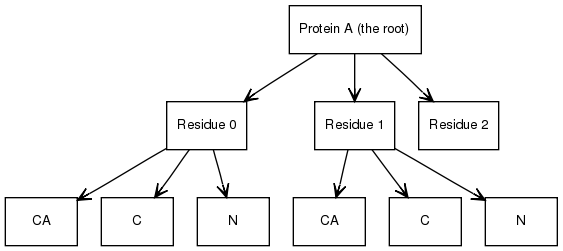
Here is a simple example with a protein with three residues. Two of the residues have atoms, where as the third is coarse grained.
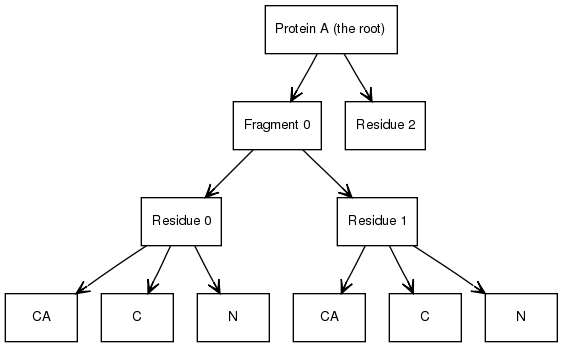
The hierarchy can be used to add extra, not-necessarily biological, structure such as domains or other convenient divisions of the molecule. For example, the protein node could have an intermediate layer between it and the first two residues:
A hierarchy can have any tree structure as long as:
The get_is_valid() method checks some of these.
A number of decorator types are associated with the Hierarchy to store the information associated with that node in the hierarchy. Examples include Residue, Atom, XYZ, Chain, XYZR, Mass, Domain etc. We provide a get_as_x() function for each such decorator which returns either X() (a null type) if the node is not a particle of type x, or an X decorator wrapping the current particle if it is.
Public Member Functions | |
| unsigned int | add_child (Hierarchy o) |
| Add a child and check that the types are appropriate. | |
| void | add_child_at (Hierarchy o, unsigned int i) |
| Add a child and check that the types are appropriate. | |
| Hierarchy | get_child (unsigned int i) const |
| HierarchiesTemp | get_children () const |
| bool | get_is_valid (bool print_info) const |
| Return true if the hierarchy is valid. | |
| Hierarchy | get_parent () const |
| Hierarchy () | |
| null constructor | |
| Hierarchy (Particle *p) | |
| Hierarchy (IMP::core::Hierarchy h) | |
| The traits must match. | |
| void | show (std::ostream &out=std::cout) const |
Methods to get associated decorators | |
We provide a number of helper methods to get associated decorators for the current node in the hierarchy. As an example, if the particle decorated by this decorator is a Residue particle, then get_as_residue() return Residue(get_particle()), if not it returns Residue(). | |
| Atom | get_as_atom () const |
| Chain | get_as_chain () const |
| Domain | get_as_domain () const |
| Fragment | get_as_fragment () const |
| Mass | get_as_mass () const |
| Residue | get_as_residue () const |
| core::XYZ | get_as_xyz () const |
| core::XYZR | get_as_xyzr () const |
Static Public Member Functions | |
| static Hierarchy | decorate_particle (Particle *p) |
| cast a particle which has the needed attributes | |
|
static const IMP::core::HierarchyTraits & | get_traits () |
| Get the molecular hierarchy HierararchyTraits. | |
| static bool | particle_is_instance (Particle *p) |
| static Hierarchy | setup_particle (Particle *p) |
Related Functions | |
| (Note that these are not member functions.) | |
| Hierarchy | create_clone (Hierarchy d) |
| Clone the Hierarchy. | |
| Hierarchy | create_clone_one (Hierarchy d) |
| Clone the node in the Hierarchy. | |
| Hierarchy | create_fragment (const HierarchiesTemp &ps) |
| Create a fragment containing the specified nodes. | |
| Hierarchy | create_protein (Model *m, double resolution, int number_of_residues, int first_residue_index=0, double volume=-1, double spring_strength=1) |
| Create a coarse grained molecule. | |
| void | destroy (Hierarchy d) |
| Delete the Hierarchy. | |
| Atom | get_atom (Residue rd, AtomType at) |
| Return a particle atom from the residue. | |
| algebra::SphereD< 3 > | get_bounding_sphere (const Hierarchy &h) |
| HierarchiesTemp | get_by_type (Hierarchy mhd, GetByType t) |
| HierarchiesTemp | get_leaves (Hierarchy h) |
| Hierarchy | get_residue (Hierarchy mhd, unsigned int index) |
| Get the residue with the specified index. | |
| Residue | get_residue (Atom d, bool nothrow=false) |
| Return the Residue containing this atom. | |
| Hierarchy | get_root (Hierarchy h) |
| Return the root of the hierarchy. | |
| void | show (Hierarchy h, std::ostream &out=std::cout) |
| Print out a molecular hierarchy. | |
PDB Reading | |
The read PDB methods produce a hierarchy that looks as follows:
Waters are currently dropped if they are ATOM records. This can be fixed. The read_pdb() functions should successfully parse all valid pdb files. It can produce warnings on files which are not valid. It will attempt to read such files, but all bets are off. When reading PDBs, PDBSelector objects can be used to choose to only process certain record types. See the class documentation for more information. When no PDB selector is supplied for reading, the NonWaterPDBSelector is used.
Set the IMP::LogLevel to IMP::VERBOSE to see details of parse errors. | |
| Hierarchies | read_multimodel_pdb (TextInput in, Model *model, const PDBSelector &selector) |
| Hierarchy | read_pdb (TextInput in, Model *model, const PDBSelector &selector, bool select_first_model=true) |
| Hierarchy | read_pdb (TextInput in, Model *model) |
PDB Writing | |
The methods to write a PDBs expects a Hierarchy that looks as follows:
All Residue particles that have a Chain particle as an ancestor are considered part of a protein, DNA or RNA, ones without are considered heterogens.
The functions produce files that are not valid PDB files, eg only ATOM/HETATM lines are printed for all Atom particles in the hierarchy. Complain if your favorite program can't read them and we might fix it. | |
| void | write_multimodel_pdb (const Hierarchies &mhd, TextOutput out) |
| void | write_pdb (const Hierarchies &mhd, TextOutput out) |
| void | write_pdb (Hierarchy mhd, TextOutput out) |
| unsigned int IMP::atom::Hierarchy::add_child | ( | Hierarchy | o | ) |
Add a child and check that the types are appropriate.
A child must have a type that is listed before the parent in the Type enum list.
| void IMP::atom::Hierarchy::add_child_at | ( | Hierarchy | o, | |
| unsigned int | i | |||
| ) |
Add a child and check that the types are appropriate.
A child must have a type that is listed before the parent in the Type enum list.
| Hierarchy IMP::atom::Hierarchy::get_child | ( | unsigned int | i | ) | const |
Get the ith child
Reimplemented from IMP::core::Hierarchy.
| bool IMP::atom::Hierarchy::get_is_valid | ( | bool | print_info | ) | const |
Return true if the hierarchy is valid.
Print information about the hierarchy if print_info is true and things are invalid.
| Hierarchy IMP::atom::Hierarchy::get_parent | ( | ) | const |
Get the parent particle.
Reimplemented from IMP::core::Hierarchy.
| static bool IMP::atom::Hierarchy::particle_is_instance | ( | Particle * | p | ) | [static] |
Check if the particle has the needed attributes for a cast to succeed
Reimplemented from IMP::Decorator.
Reimplemented in IMP::atom::Atom, and IMP::atom::CHARMMAtom.
Create a Hierarchy of level t by adding the needed attributes.
| void IMP::atom::Hierarchy::show | ( | std::ostream & | out = std::cout |
) | const |
Write information about this decorator to out.
| Hierarchy create_protein | ( | Model * | m, | |
| double | resolution, | |||
| int | number_of_residues, | |||
| int | first_residue_index = 0, |
|||
| double | volume = -1, |
|||
| double | spring_strength = 1 | |||
| ) | [related] |
Create a coarse grained molecule.
The coarse grained model is created with a number of spheres based on the resolution and the volume. If the volume is not provided it is estimated based on the number of residues. The protein is created as a molecular hierarchy rooted at p. The leaves are Domain particles with appropriate residue indexes stored and are XYZR particles.
Volume is, as usual, in cubic anstroms.
Currently the function creates a set of balls with radii no greater than resolution which overlap by 20% and have a volume of their union equal to the passed volume. The balls are held together by a ConnectivityRestraint with the given spring constant.
The coordinates of the balls defining the protein are optimized by default, and have garbage coordinate values.
| void destroy | ( | Hierarchy | d | ) | [related] |
Return a particle atom from the residue.
The residue must be part of a molecular hierarchy.
| algebra::SphereD< 3 > get_bounding_sphere | ( | const Hierarchy & | h | ) | [related] |
See get_bounding_box() for more details.
| HierarchiesTemp get_leaves | ( | Hierarchy | h | ) | [related] |
Return the Residue containing this atom.
The atom must be part of a molecular hierarchy.
| ValueException | if no residue is found, unless nothrow is true. |
Read a all the molecules in the first model of the pdb file.