Data and code for Integrative Structure Determination using data from point mutant epistatic miniarray profile (pE-MAP).
pE-Map data
The clustered pE-MAP for the histones and correlation maps are available in the supplementary data of [https://science.sciencemag.org/content/370/6522/eaaz4910.abstract].
The yeast RNAPII pE-MAP is available in the supplementary materials of [https://www.sciencedirect.com/science/article/pii/S0092867413009380].
The bacterial RNAP point mutation data are available in the supplementary materials of [https://www.biorxiv.org/content/10.1101/2020.06.16.155770v1.abstract].
pE-Map restraint
Four files are included in the restraint directory:
- update_pemap_imp.sh
- include/PEMAPRestraint.h
- src/PEMAPRestraint.cpp
- restraint/pyext/src/pemap.py
Use the update_pemap_imp.sh file to copy the pEMAP restraint files into the IMP directory. The update_pemap_imp.sh file needs to be edited to indicate the path your local IMP directory. Additionally, edit the file modules/isd/pyext/swig.i-in to add the following lines:
IMP_SWIG_OBJECT(IMP::isd, PEMAPRestraint, PEMAPRestraints);
%include "IMP/isd/PEMAPRestraint.h"
Make sure to re-compile the code after adding the files.
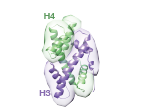
Modeling of the H3-H4 dimer
Data for simulations (directory modeling_histones/data)
Data used for modeling includes:
-
Script and alignments to generate comparative models.
-
Comparative models of H3 and H4
-
Processed pE-MAP data file containing the pairs of residues to which the pE-MAP distance restraints will be applied. The format of these files is:
protein1 protein2 residue1 residue2 MIC_value distance_in_xray_structure (if known)
- Python scripts to shuffle and resample the pE-MAP data files
Running the simulations (directory modeling_histones/modeling)
-
top_his_comp_models.dat: Topology file containing the representation of the h3-h4 system. Each protein has presented as a rigid body. Histones tails are not considering for modeling. -
mod_pemap_histones.py: PMI modeling scripts for running the production simulations. The search for good-scoring models relied on Replica Exchange Gibbs sampling, based on the Metropolis Monte Carlo (MC) algorithm. We recommend producing at least 2,500,000 models from 50 independent runs, each starting from a different initial conformation of H3-H4 dimer to have proper statistics.
Analysis of simulations (directory modeling_histones/analysis)
The analysis uses the PMI_analysis module in: https://github.com/salilab/PMI_analysis
-
run_analysis_trajectories.py: -
run_extract_models.py: Get the rmf3 files for a random sampled of 30,000 structures in the ensemble. These structures are split into two sets (sample_A, sample_B) -
run_clustering.sh:
Modeling of RNAP II
Modeling of Bacterial RNAP II
Other files
Information
Author(s): Ignacia Echeverria, Hannes Braberg
Date: November 24th, 2020
License: LGPL. This library is free software; you can redistribute it and/or modify it under the terms of the GNU Lesser General Public License as published by the Free Software Foundation; either version 2 of the License, or (at your option) any later version.
Testable: Yes
Parallelizeable: Yes
Publications: Braberg, H., Echeverria, I., Bohn, S., Cimermancic, P., Shiver, A., Alexander, R., Xu, J., Shales, M., Dronamraju, R., Jiang, S. and Dwivedi, G., Bogdanoff D., Chaung K. K., Hüttenhain R., Wang S., Mavor D., Pellarin R., Schneidman D., Bader J. S., Fraser J. S., Morris J., Haber J. E., Strahl B. D., Gross C. A., Dai J., Boeke J. D., Sali A., Krogan N. J. 2020. Genetic interaction mapping informs integrative structure determination of protein complexes. Science, 370(6522). [https://science.sciencemag.org/content/370/6522/eaaz4910.abstract]



 Download files
Download files Warning: these files have not yet been verified to work with the latest version of IMP. We will update this page when they have been.
The files are also available at
Warning: these files have not yet been verified to work with the latest version of IMP. We will update this page when they have been.
The files are also available at  To install the software needed to reproduce this system with the
To install the software needed to reproduce this system with the
 To set up the environment on the UCSF Wynton cluster to run
this system, run:
To set up the environment on the UCSF Wynton cluster to run
this system, run: