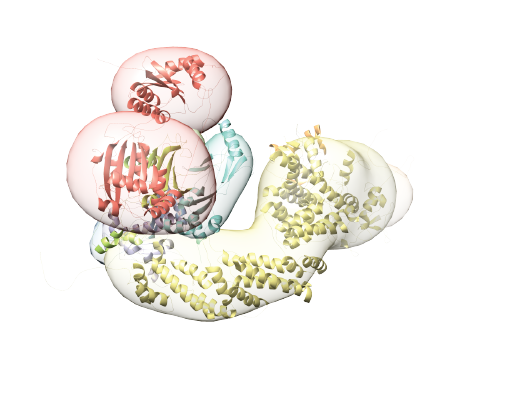
Integrative structure determination of the A3G-VifHIV-1-CRL5-CBFβ complex.
Summary
Integrative structure determination of the A3G-CRL5-Vif-CBFβ complex based on the 132 DSSO cross-links, atomic structures of the components, and previously published mutagenesis data. Preliminary integrative modeling represented the CRL5-Vif-CBFβ complex by a single rigid body defined by atomic structure of the Vif-CBFβ-Cul5N-term-EloB-EloC subcomplex (PDB access code: 4N9F) and comparative models of the full length Cul5 and Rbx2 (rigid representation). Following, to characterize the flexibility of the complex, we performed integrative structure modeling of the A3G-CRL5-Vif-CBFβ complex with a modified representation that allowed for relaxation in the configuration of the VCBC subcomplex subunits and for alternative conformations of Cul5 and A3G subunits (flexible representation). The structure was cross-validated based on the data used for modeling and previously published structural, biochemical, and functional data not used for modeling.
List of files and directories:
dataAll data used for integrative modeling, includingMODELLERscripts to generate the comparative models of the components, atomic structures of the components, and crosslinking data.scriptsTopology files for the rigid (top_A3G_CRL5_3rb.dat)and flexible (top_A3G_CRL5_flex.dat) representations and PMI modeling scripts for the rigid (mod_A3G_Vif_CRL5_rigid.py) and flexible (mod_A3G_Vif_CRL5_flexible.py) representations.analysisScripts to analyze the simulationsresultsAll the relevant results for the rigid and flexible representations.SI_tableScripts to generate a table summarizing the integrative modeling protocols.utilsTemplate and code to generate the Supporting information table summarizing the integrative modeling protocol.
Information
Author (s): Ignacia Echeverria
License: CC BY-SA 4.0 This work is licensed under the Creative Commons Attribution-ShareAlike 4.0 International License.
Publications: Characterization of a A3G-VifHIV-1-CRL5-CBFβ structure using a cross-linking mass spectrometry pipeline for integrative modeling of host-pathogen complexes. Robyn M. Kaake, Ignacia Echeverria, Seung Joong Kim, John Von Dollen, Nicholas M. Chesarino, Yuqing Feng, Clinton Yu, Hai Ta, Linda Chelico, Lan Huang, John Gross, Andrej Sali, Nevan J. Krogan, DOI:https://doi.org/10.1016/j.mcpro.2021.100132



 Download files
Download files Warning: these files have not yet been verified to work with the latest version of IMP. We will update this page when they have been.
The files are also available at
Warning: these files have not yet been verified to work with the latest version of IMP. We will update this page when they have been.
The files are also available at  To install the software needed to reproduce this system with the
To install the software needed to reproduce this system with the
 To set up the environment on the UCSF Wynton cluster to run
this system, run:
To set up the environment on the UCSF Wynton cluster to run
this system, run: