IMP.
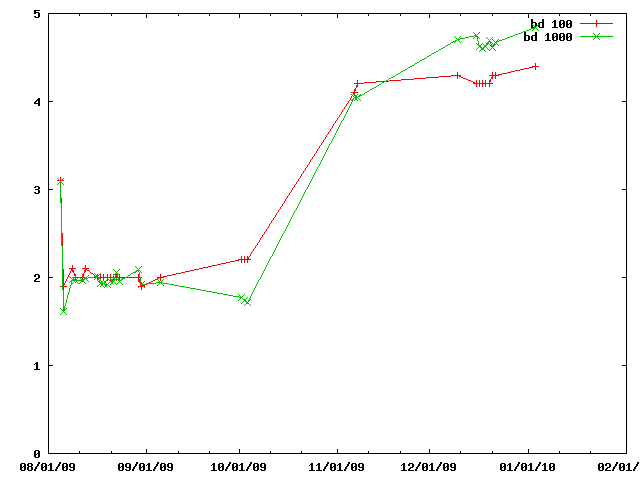
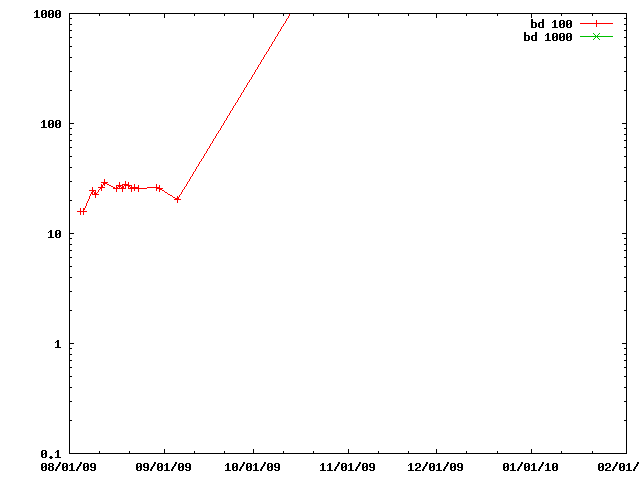
A simple test modifying the particles and (for the 'score' entries) evaluating a simple, incremental, score.
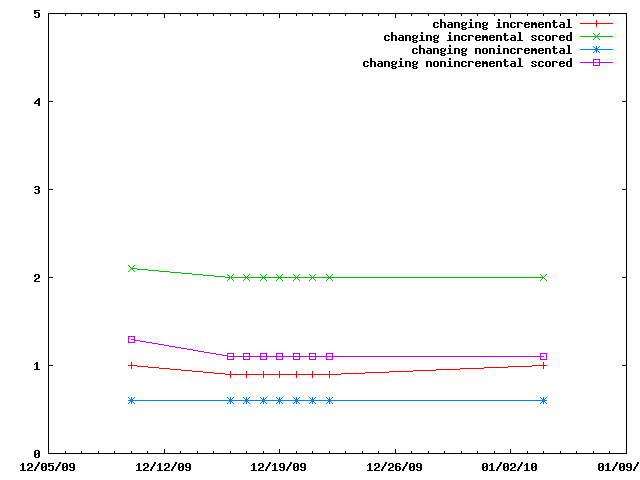
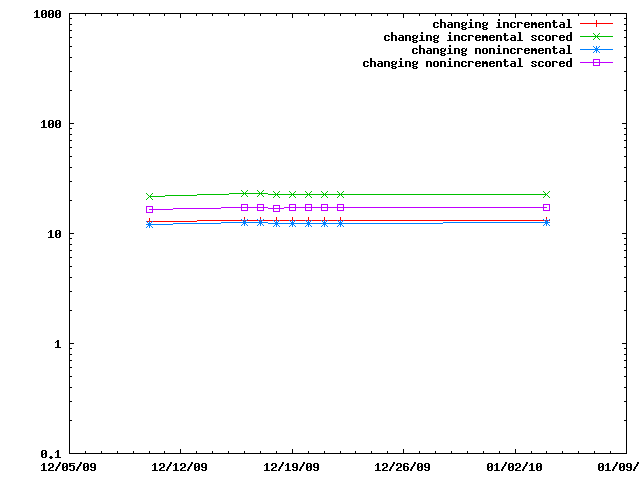
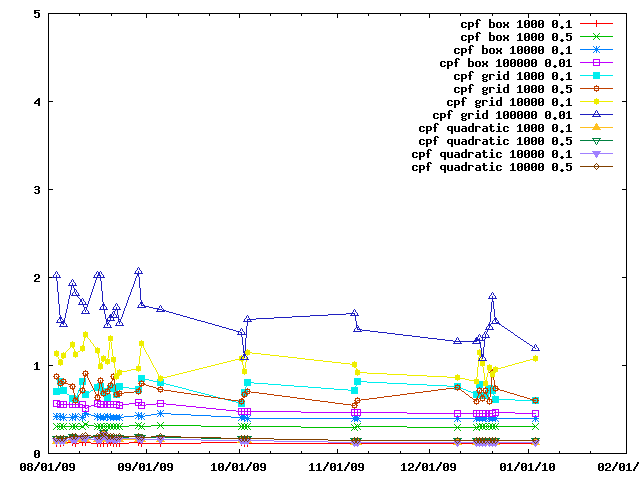
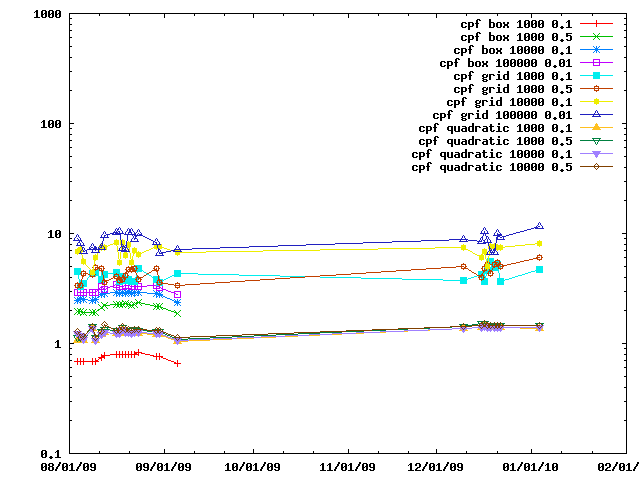
The various IMP::core::ClosePairFinder implementations are benchmarked on random sets of particles. The benchmarks for the quadratic one are listed as taking 10x less time than they did.
A benchmark of the various close pairs finders in the context of a simulation. The benchmarks for the quadratic one are listed as taking 10x less time than they did.
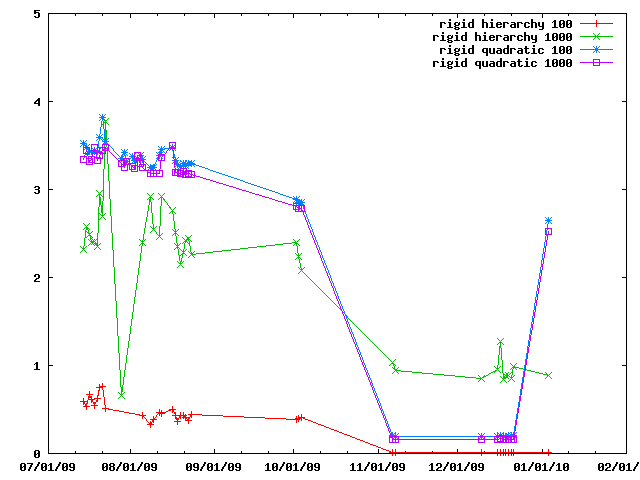
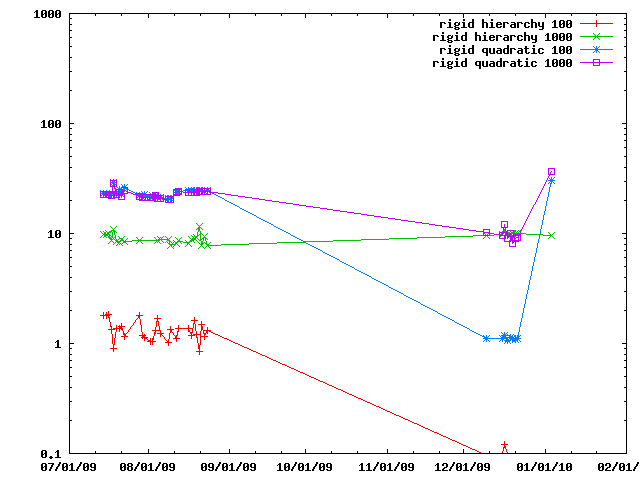
A special close pair finder is provided for rigid bodies, name IMP::core::RigidClosePairsFinder. Here it is compared against using all the member particles.
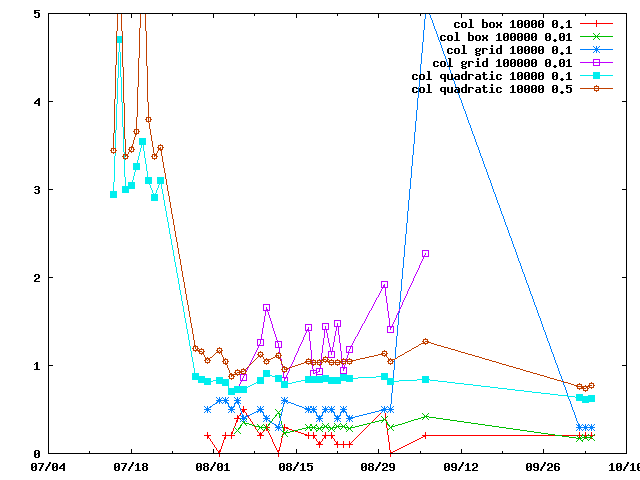
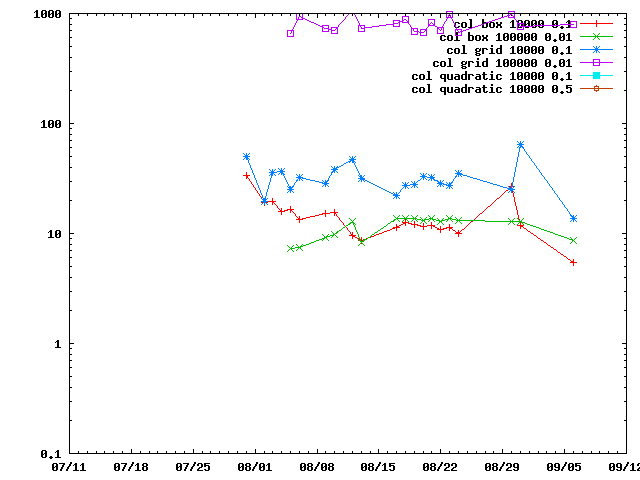
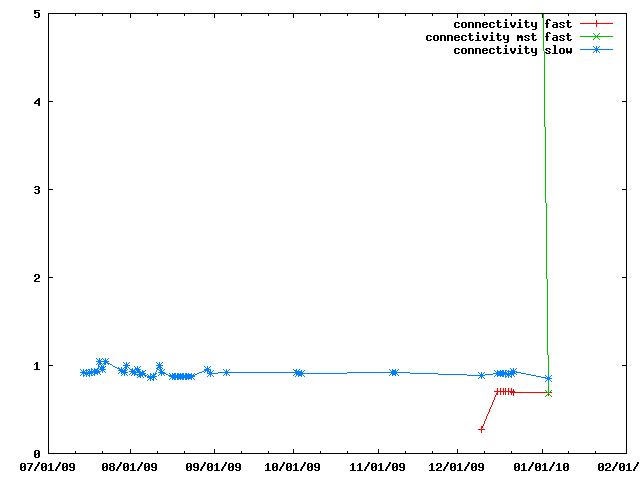
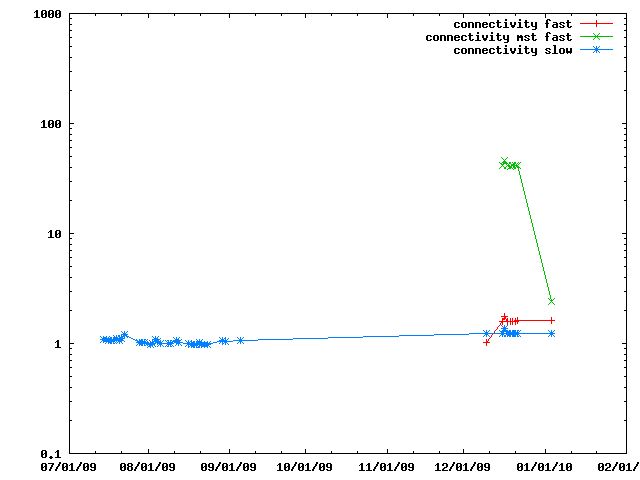
There are several ways of doing connectivity restraints in IMP: IMP::misc::ConnectedPairContainer and IMP::core::ConnectivityRestraint. They are benchmarked here.
The next benchmark looks at evaluating restraints on various sorts of containers, namely IMP::core::ListSingletonContainer and IMP::core::SingletonContainerSet.
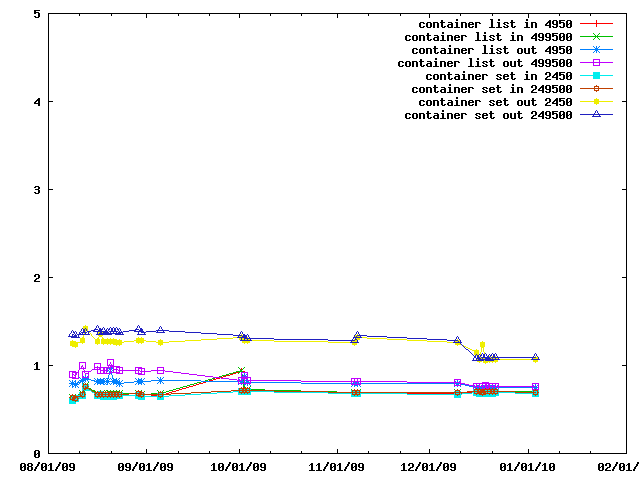
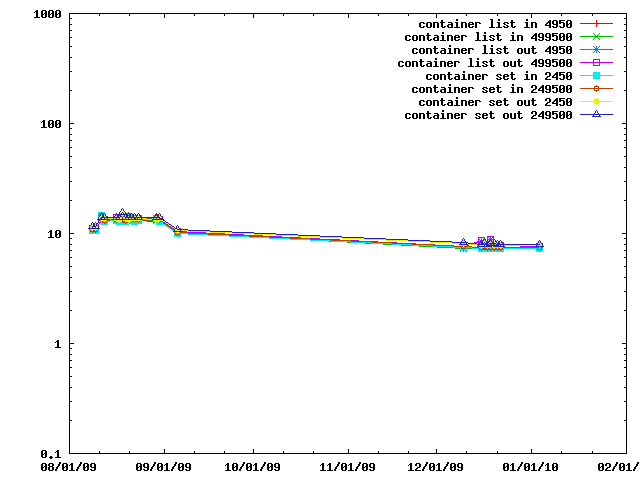
When peforming lots of operations on a set of cartesian coordinates and is faster to copy them into a IMP::algebra::Vector3Ds than to access them in the IMP::core::XYZ decorator each time. This benchmark gives an idea of the speed tradeoffs involved.
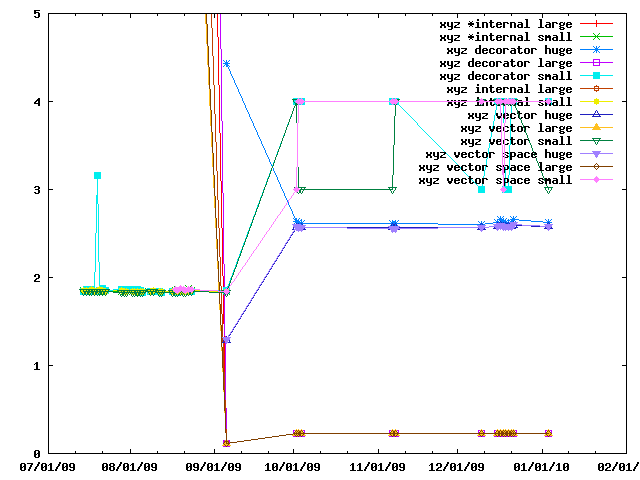
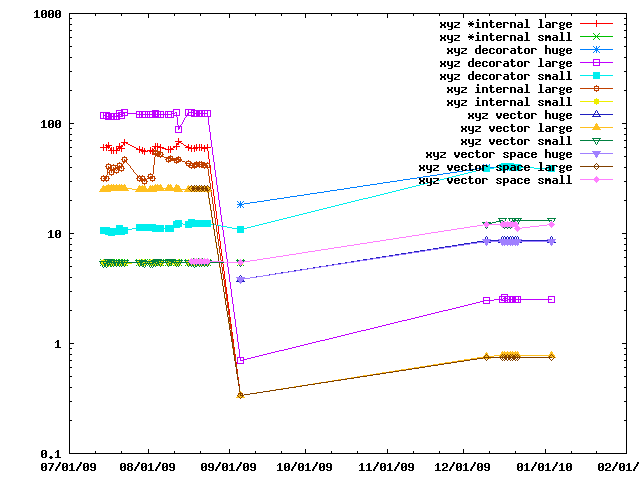
IMP and how to apply them to biological problems.
Functions | |
| std::string | get_data_path (std::string file_name) |
| Return the path to installed data for this module. | |
| std::string | get_example_path (std::string file_name) |
| Return the path to installed example data for this module. | |
| std::string | get_module_name () |
| const VersionInfo & | get_module_version_info () |
| void | report (std::string name, double value, double check) |
| Report a benchmark result in a standard way. | |
| std::string IMP::benchmark::get_data_path | ( | std::string | file_name | ) |
Return the path to installed data for this module.
Each module has its own data directory, so be sure to use the version of this function in the correct module. To read the data file "data_library" that was placed in the data directory of module "mymodule", do something like
std::ifstream in(IMP::mymodule::get_data_path("data_library"));
IMP is installed or used via the tools/imppy.sh script.
| std::string IMP::benchmark::get_example_path | ( | std::string | file_name | ) |
Return the path to installed example data for this module.
Each module has its own example directory, so be sure to use the version of this function in the correct module. For example to read the file example_protein.pdb located in the examples directory of the IMP::atom module, do
IMP::atom::read_pdb(IMP::atom::get_example_path("example_protein.pdb", model));
IMP is installed or used via the tools/imppy.sh script.
| void IMP::benchmark::report | ( | std::string | name, | |
| double | value, | |||
| double | check | |||
| ) |
Report a benchmark result in a standard way.
The last value is a check value which can be used to make sure the computations were semi-ok.