IMP is a tool to generate structural models based on multiple sources of experimental data. A detailed explanation of IMP and integrative modeling is given here
- Show the basic capabilities of IMP
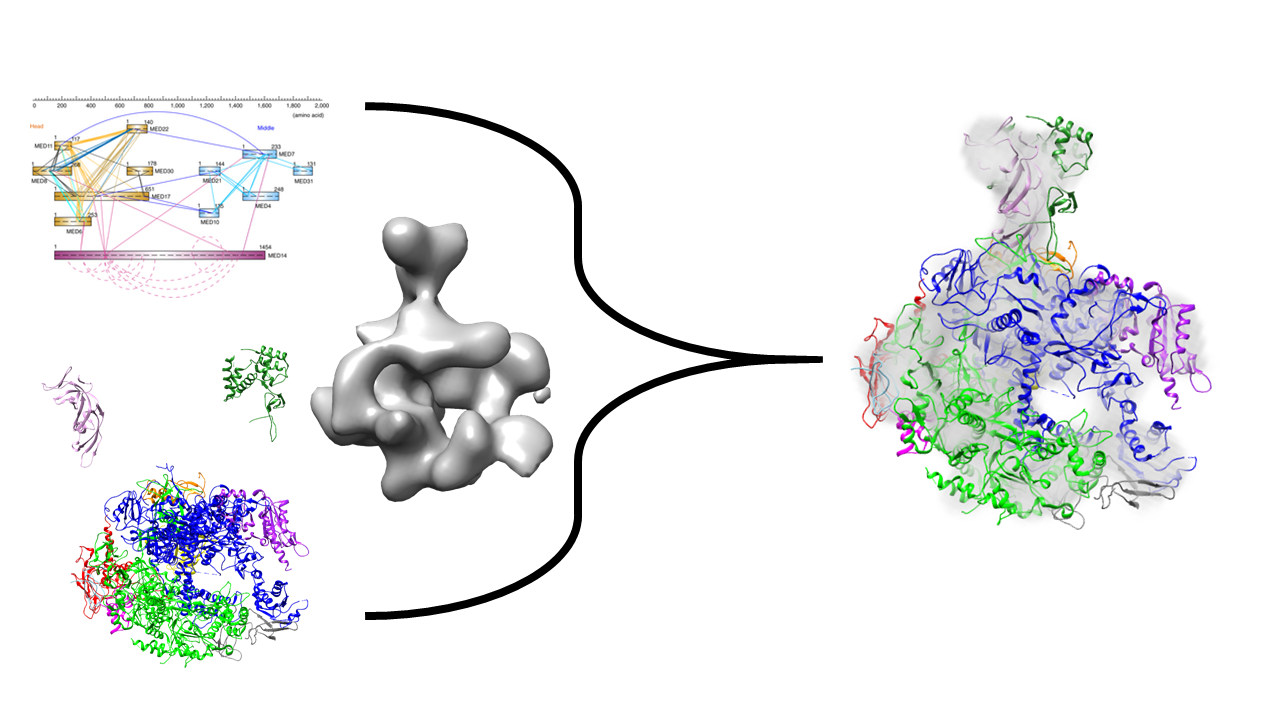
- Sample models of RNA Polymerase II using Electron Microscopy density and chemical crosslinks
- Visualize these models and analyze them for accuracy and precision
See the IMP website for the tutorial text.


 Download files
Download files Verified to work with the
Verified to work with the  To install the software needed to reproduce this system with the
To install the software needed to reproduce this system with the
 To set up the environment on the UCSF Wynton cluster to run
this system, run:
To set up the environment on the UCSF Wynton cluster to run
this system, run: