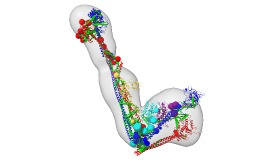
This repository contains Python scripts and associated input data and output results for constructing the integrative model of the ATP-bound five-subunit yeast Smc5/6-Nse2/5/6 complex, using IMP, (develop-255ae6c), PMI and MODELLER.
The input data used for this work is:
- Crystal structure of the a part of the coiled-coil arm of Smc5 in complex with Nse2 (PDB ID: 3htk).
- Comparative models of head and hinge regions of Smc5 and Smc6.
- Molecular-mechanics based (energy-minimized) coiled-coil models of Smc5 and Smc6 arm regions.
- Cryo-EM derived structure of Nse5/6 subcomplex (at 3 angstroms resolution)
- 268 unique DSSO and 67 unique CDI crosslinks.
First, comparative models of the different parts of Smc5/6 as mentioned above, are constructed using both MODELLER and the SWISS-MODEL webserver. Next, parametric models of the coiled-coil regions of Smc5/6 arms are designed using the protein design software package ISAMBARD. Finally, models of these different components are put together and the crosslink information is used to create spatial restraints and derive the overall model of the five subunit complex using IMP. The modeling protocol will work with a default build of IMP, but for most effective sampling, IMP should be built with MPI so that replica exchange can be used.
-
smc56_domain_definitions.txtcontains the break-down of different domains of Smc5/6 used to designate / design rigid bodies for integrative modeling. -
datacontains all input data used in integrative modeling.-
Crosslink data in the folder
data/xl -
Pdb files (comparative models, coiled-coil models, crystal structures) in
data/pdb -
Sequences in
smc56_nse256.fasta.txt -
System topology description for PMI to process in
topology.txt.
-
-
non_cc_comparative_modelingcontains all comparative models for non-coiled-coil regions of Smc5/6, i.e. the head and hinge regions.-
Hinge region models generated using SWISS-MODEL are kept in folders of the corresponding name and suffixed SWISSMODEL.
-
Comparative models of the combined Smc5/6 head region is generated using MODELLER and kept in
non_cc_comparative_modeling/smc56_head_MODELLER. -
The final set of comparative models are kept in
non_cc_comparative_modeling/best_models. -
.pngfiles of comparative model structures rendered using UCSF Chimerax are kept innon_cc_comparative_modeling/images. -
Different steps of the comparative modeling protocol for hinge and head regions are detailed in the jupyter notebook
non_cc_comparative_modeling/modeling.ipynb.
-
-
cc_parametric_modelingcontains scripts and results for coiled-coil models designed using ISAMBARD.- Coiled coils designed for Smc5/6 are kept in directories
cc_parametric_modeling/smc5andcc_parametric_modeling/smc6respectively. Each directory contains separate folders for coiled-coils from the head-adjacent (cc-head), hinge-adjacent(cc-hinge) and middle (cc-mid-1andcc-mid-2) parts of the Smc5/6 arm. - The final set of coiled coil models are kept in
cc_parametric_modeling/best_models. - .png files of coiled coil models are kept in
cc_parametric_modeling/images. - Python script and utility files for searching coiled-coil design parameters are also kept in this directory.
- Different steps of the coiled coil design protocol are detailed in the jupyter notebook
cc_parametric_modeling/modeling.ipynb.
- Coiled coils designed for Smc5/6 are kept in directories
-
integrative_modelingcontains (Python and Bash) scripts and results for building integrative models of the complex and performing structural calculations on these models.-
structural_clusteringcontains results produced by the model validation pipeline (good scoring model rmf files, model precision, localilzation densities). -
structural_calculationscontains crosslink satisfaction results, and the Smc5/6 arm length and elbow angle.
-
-
rendering_scriptscontains python scripts to render convert RMF files to PDB files and for producing the 2D crosslinked schematic in Fig. 2 of the manuscript.
First, non-comparative models are generated following the steps outlined in the jupyter notebook non_cc_comparative_modeling/modeling.ipynb. The model of Smc5/6 head regions is generated by running the MODELLER script non_cc_comparative_modeling/smc56_head_MODELLER/modeling.py as:
cd non_cc_comparative_modeling/smc56_head_MODELLER
python modeling.pyThis will produce all output files and a log file (log.txt) in this directory. To keep things clean, all of this output has been stored in a separate subfolder non_cc_comparative_modeling/smc56_head_MODELLER/output. The final models produced from this step are all stored in non_cc_comparative_modeling/best_models and should be copied over to data/pdb to be used later in integrative modeling.
Next, coiled-coil models are designed by searching in a parameter space spanned by geometrical descriptors of coiled-coil structures using the ISAMBARD software package. The motivation and all necessary steps are outlined in the notebook cc_parameteric_modeling/modeling.ipynb and demonstrated in detail for a specific example of the Smc5 coiled coil region adjacent to the head region. The protocol is similar for other coiled coil segments. The final models produced from this step are all stored in cc_parametric_modeling/best_models and should be copied over to data/pdb to be used later in integrative modeling.
At this stage, PDB files corresponding to different rigid bodies in the complex must be ready and kept in data/pdb. The rigid body definitions can be found in the PMI topology file data/topology.txt. All python scripts in this folder can be run with a -h flag, i.e. python <script name> -h to see its usage, inputs, expected outputs, etc.
-
First use
integrative_modeling/modeling.pyto build the complex representation, add the spatial restraints and performs conformational sampling. It has a test mode (use flag-t) to run a very short number of Monte Carlo iterations and verify that everything works. Keep results of independent runs using this script in directories calledrun_1, run_2, etc. Do at least 50 independent runs (you may need a computing cluster resource) for realistic results. Sample usage (that produces output in a directory calledrun_1):cd integrative_modeling mkdir run_1 cd run_1 python ../modeling.py -d ../../data
-
Next, use
integrative_modeling/analyse_score_distribution.pyto select equilibrated models and filter them according to their scores (i.e. restraint satisfaction). This calculates restraint satisfactions for all independent runs and also clusters the models based on their scores. Keep all output generated in a directory calledanalys. Sample usage:cd integrative_modeling python analyse_score_distributions.py -i . -o analys -np 20
-
Next, For structural clustering of models, extract a random subsample of good scoring models from the output produced by the score analysis pipeline in the previous step. To do that, run
integrative_modeling/get_good_scoring_models.pyas:cd integrative_modeling python get_good_scoring_models -a analys -t . -o structural_clustering -np 20
This produces all the good scoring models, but partitions them into two sets called
sample_A_models.rmf3andsample_B_models.rmf3, kept in thestructural_clusteringdirectory. The two sub-samples are necessary for running statistical validation tests later, but you can go ahead and concatenate them to produce a single file containing (~30,000) good scoring models. Call thisgood_scoring_models.rmf3.cd integrative_modeling/structural_clustering rmf_cat sample_A_models.rmf3 sample_B_models.rmf3 good_scoring_models.rmf3where,
rmf_catis an executable utility for combining frames from different RMF files and ships with IMP. This produces the fileintegrative_modeling/structural_clustering/good_scoring_models.rmf3. -
Next, run the executable
model_precision_job.shto go through a statistical structural precision analysis pipeline to calculate sampling precision, model precision and cluster the good scoring models kept ingood_scoring_models.rmf3according to RMSD. It turns out that using default settings, the clustering is too stringent, i.e. > 100 clusters are produced. To make the clustering smoother, edit the shell script by removing the comment before# -s -ct 50and rerun clustering. So, the first run is to extract the sampling precision only. Make different directories for these two cases. Recommended directory names aresp_autofor calculating sampling precision (with automatic / default settings) and thensp_50to cluster at 50 Å threshold. For calculating only sampling precision:cd integrative_modeling/structural_clustering mkdir sp_auto cd sp_auto cp ../../model_precision_job.sh ./ cp ../../density_ranges.txt ./ ./model_precision_job.sh
Only when this run has finished, rerun at 50 Å threshold.
cd integrative_modeling/structural_clustering mkdir sp_50 cd sp_50 cp ../sp_auto/model_precision_job.sh ./Make the necessary tweak to this version of
model_precision_job.sh. Then:cp ../sp_auto/Distance_matrix.npy ./ cp ../sp_auto/density_ranges.txt ./ ./model_precision_job.sh
Note that in each case, the file
density_ranges.txtneeds to be present in the directory from where the shell script is run. This file contains subunit definitions for which 3D localization densities are calculated. For the re-run, a pairwise distance matrix of alignment RMSD among all modelsDistance_matrix.npyis also required; just copy it fromsp_auto. This produces the cluster centroids and localization densities in directories calledsp_50/cluster.0,sp_50/cluster.1and so on, and model precision and other statistics for each cluster insp_50.Note that in this repository, only selected results from
structural_clusteringandanalyshave been retained.
The integrative model is represented not only by the centroid, but the ensemble of all good scoring models in the most populated cluster. To extract structural metrics like Smc5/6 arm length, etc, the calculation should be repeated for all models in the top-cluster so as to produce a mean and an error-bar for that metric. All of these results are kept in integrative_modeling/structural_calculations. To calculate crosslink satisfaction (and store the results in a folder called structural_calculations/xl_satisfaction):
cd integrative_modeling/structural_calculations
python ../get_XL_satisfaction.py -r ../structural_clustering/good_scoring_models.rmf3 -c 0 -m ../structural_clustering/sp_50/cluster.0.all.txt -xl ../../data/xl/xl_all.csv -o ./xl_satisfaction/To calculate Smc5/6 arm geometry related metrics (and store it in a file called structural_calculations/Smc56_arm_geometry.txt):
cd integrative_modeling/structural_calculations
python ../get_Smc56_arm_geometry.py -r ../structural_clustering/good_scoring_models.rmf3 -c 0 -m ../structural_clustering/sp_50/cluster.0.all.txt -o .Author(s): Tanmoy Sanyal
Date: April 13, 2021
License: CC BY-SA 4.0 This work is licensed under the Creative Commons Attribution-ShareAlike 4.0 International License.
Testable: Yes.
Parallelizable: Yes
Publications: You Yu, Shibai Li, Zheng Ser, Tanmoy Sanyal, Koyi Choi, Bingbing Wan, Huihui Kuang, Andrej Sali, Alex Kentsis, Dinshaw J. Patel and Xiaolan Zhao, Integrative analysis reveals unique structure and function features of the Smc5/6 complex, bioRxiv (2020)



 Download files
Download files Verified to work with the
Verified to work with the  To install the software needed to reproduce this system with the
To install the software needed to reproduce this system with the
 To set up the environment on the UCSF Wynton cluster to run
this system, run:
To set up the environment on the UCSF Wynton cluster to run
this system, run:
