 |
|
IMP Tutorial
|
In this tutorial we will introduce the procedure used to deposit integrative modeling studies in the PDB-Dev database in mmCIF format.
We will demonstrate the procedure using IMP and its PMI module, but the database will accept integrative models from any software, as long as they are compliant mmCIF files (e.g. there are several HADDOCK and Rosetta models already in PDB-Dev).
This tutorial can be followed in several ways:
rnapolii/modeling/deposition.ipynb.PDB-Dev is a database run by the wwPDB. It is specifically for the deposition of integrative models, i.e. models generated using more than one source of experimental data. (Models that use only one experiment generally go in PDB; models that use no experimental information - theoretical models - go in ModelArchive).
wwPDB already uses the mmCIF file format for X-ray structures, and has a formal data model (PDBx) to describe these structures. The format is extensible; extension dictionaries exist to describe NMR, SAS, EM data, etc. For integrative models, we use PDBx plus an "integrative/hybrid methods" (IHM) extension dictionary. This supports coarse-grained structures, multiple input experimental data sources, multiple states, multiple scales, and ensembles related by time or other order.
A frequently asked question is whether mmCIF files can be generated directly from an IMP::Model or an RMF file. This generally isn't possible because RMF and IMP::Model are designed to store one or more output models, where each model is a set of coordinates for a single conformation of the system being studied. A deposition, on the other hand, aims to cover a complete modeling study, and should capture the entire ensemble of output models, all of the input data needed to reproduce the modeling, and quality metrics such as the precision of the ensemble and the degree to which it fits the output data. A deposition is also designed to be visualized, and so may contain additional data not used in the modeling itself, such as preset colors or views to match figures in the publication or highlight regions of interest, and more human-descriptive names for parts of the system. Thus, deposition is largely a data-gathering exercise, and benefits from a modeling study being tidy and well organized (for example, by storing it in a GitHub repository) so that data is easy to find and track to its source.
The file format used by PDB for deposition of integrative modeling studies is the same mmCIF format that is used for crystal structures, with extensions (the Integrative/Hybrid Methods (IHM) dictionary) to support coarse-grained structures, multiple input experimental data sources, multiple states, multiple scales, and ensembles related by time or other order. Currently, models that are compliant with the IHM dictionary can be deposited in the PDB-Dev database.
mmCIF is a text format with a well-defined syntax, so in principle files could be generated by hand or with simple scripts. However, it is generally easier to use the existing python-ihm library. This stores the same data as in an mmCIF file, but represents it as a set of Python classes, so it is easier to manipulate. IMP includes a copy of the python-ihm library, and uses it internally to read and write mmCIF files.
There are two main approaches to generating an mmCIF file for deposition:
The first approach offers the greatest flexibility for custom protocols or for modeling outside of IMP itself, but does require everything to be done manually. For a good example of this approach, see the modeling of Nup133. (One example of the tradeoff between flexibility and manual coding: the cross-links used in the study are stored in a plain text file so a custom Python class had to be written to parse them.)
The second approach will be explored in this tutorial.
ProtocolOutput is designed to be attached to a top-level PMI object (usually IMP::pmi::topology::System). Then, as the script is run, it will capture all of the information about the modeling study, in an ihm.System object. Additional information not in the modeling script itself, such as the resulting publication, can be added using the python-ihm API.
We will demonstrate ProtocolOutput by using it to make a deposition of the RNA Pol II stalk modeling, as covered in the introductory PMI tutorial. First, download the files for this tutorial by using the "Clone or download" link at the tutorial's GitHub page.
We will then modify the modeling.py script from the introductory tutorial, calling it deposition.py in the rnapolii/modeling subdirectory, to capture the study as mmCIF.
The first modification to deposition.py is to import the PMI mmCIF and python-ihm Python modules:
Next, as soon as the top-level IMP::pmi::topology::System object is created, we attach a ProtocolOutput object (BuildSystem contains a system member):
Note that the ProtocolOutput object po simply wraps an ihm.System object as po.system. We can then customize the ihm.System by setting a human-readable title and adding a citation (here we use ihm.Citation.from_pubmed_id, which looks up a citation by PubMed ID - this particular PubMed ID is actually for the previously-published modeling of the Nup84 complex).
As it is generally undesirable to rerun the entire modeling just to generate an mmCIF file, we can save a lot of time when we do the replica exchange by turning on ReplicaExchange's test_mode, which skips the actual Monte Carlo simulation (so we use the previously-generated trajectory):
Once we're done with our PMI protocol, we call the ProtocolOutput.finalize method to collect all of the information about the integrative modeling protocol in ihm.System:
Note that the ihm.System object, stored as the system attribute of ProtocolOutput, contains a full description of the system:
Note that python-ihm only knows about three restraints, the two crosslinking restraints and the one EM density restraint, not the connectivity or excluded volume. This is because PDB considers that all valid structures satisfy connectivity and excluded volume by definition.
At this point we have basic information which we can write out to an mmCIF file. Let's do that to get an idea of what the file looks like:
mmCIF is a simple text format, so we can look at initial.cif in any text editor. Some simple data are stored in straightforward key:value pairs, e.g.
More complex data can be stored as a table using a loop construct. This lists the fields (headings for the columns) after which the data (as rows) follow, e.g.
Missing data are represented by ? if they are unknown or . if they are deliberately omitted (for example if they don't make sense here).
In each case both the categories (struct, software) and the data items (title, pdbx_ordinal, etc.) are defined by PDB, in the PDBx dictionary. This is the same dictionary used for regular PDB entries (e.g. crystal structures). Categories prefixed with ihm are in the IHM dictionary, which is specific to integrative models.
Integrative modeling draws on datasets from a variety of sources, so for a complete deposition all of this data need to be available. python-ihm provides an ihm.dataset module to describe various types of dataset, such as input experimental data, auxiliary output files (such as localization densities), or workflow files (such as Python scripts for modeling or visualization). For example, each restraint has an associated dataset:
The data are not placed directly in the mmCIF file - rather, the file contains links (using the ihm.location module). These links can be:
Database identifiers are preferable because the databases are curated by domain experts and include domain-specific information, and the files are in standard formats. ProtocolOutput will attempt to use these where possible. When a file is used for the modeling which cannot be tracked back to a database, ProtocolOutput will include its path (relative to that of the mmCIF file). For example, in this case the cross-links used are stored in simple CSV files and are linked as such:
In addition, the Python script itself is linked from the mmCIF file. Such local paths won't be available to end users, so for deposition we need to replace these paths with database IDs or DOIs (more on that later).
Datasets all support a simple hierarchy, where one dataset can be derived from another. In this case the EM restraint uses a locally-available GMM file, but it is derived from a density map which is stored in EMDB (as EMD-1883). ProtocolOutput is able to read (using the ihm.metadata module) metadata in both the GMM file and .mrc file to automatically determine this relationship:
In cases where ProtocolOutput is not able to determine the parent of a dataset, we can create a dataset manually and add it to d.parents, either directly or using the add_primary method. For example, see the modeling of the NPC where we linked our EM restraint back to the parent (the density map in EMDB) and grandparent (cryo-ET tilt series in EMPIAR from which the density map was constructed).
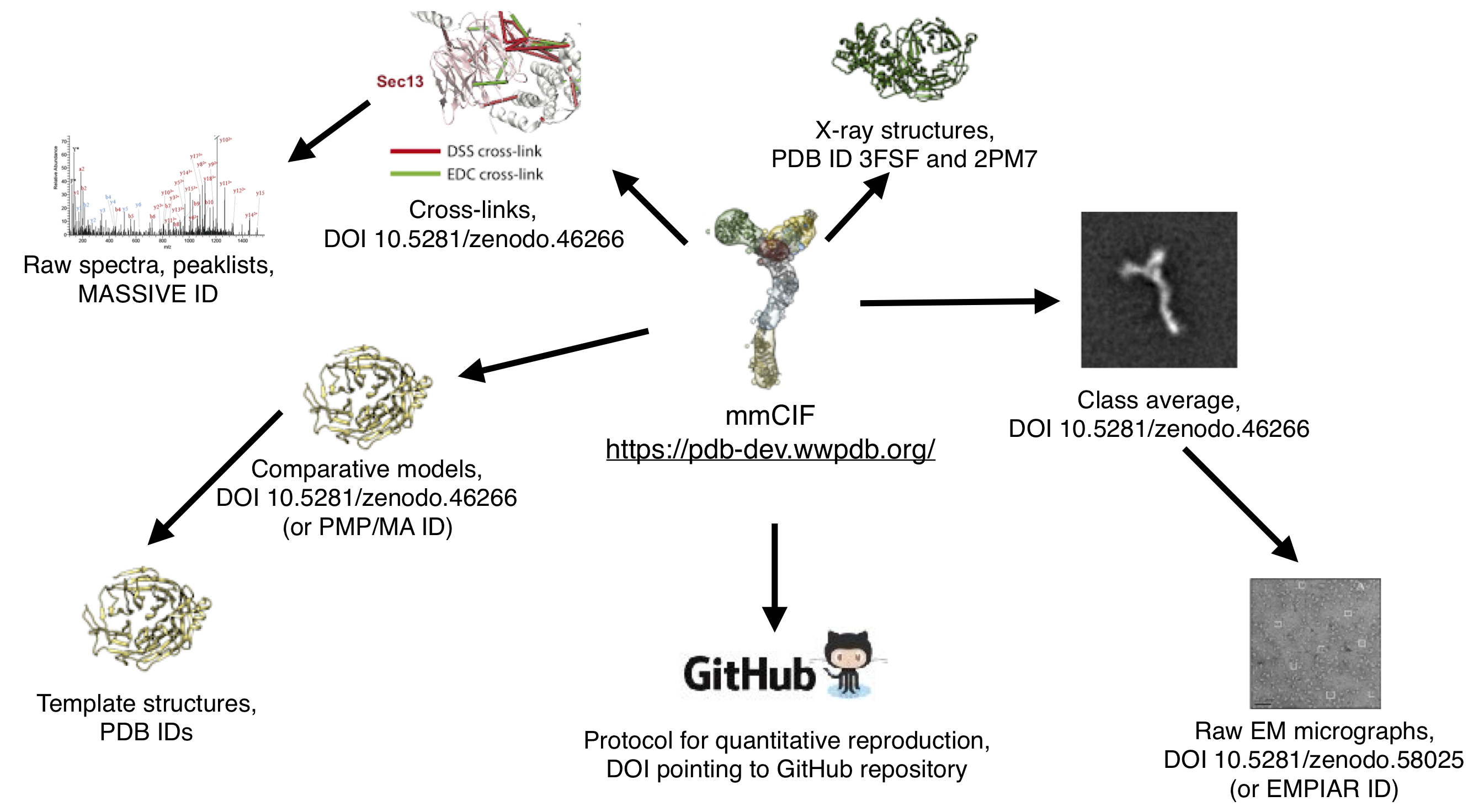
As a further example of linkage, see the links in the previously-published modeling of the Nup84 complex below. The mmCIF file links to the data directly used in the modeling (cross-links, crystal structures, electron microscopy class averages, comparative models, and Python scripts) via database IDs or DOIs. Furthermore, where available links are provided from this often-processed data back to the original data, such as templates for comparative models, mass spectometry spectra for cross-links, or micrographs for class averages:
ProtocolOutput will look at all input files to try to extract as much metadata as possible. As described above this is used to look up database identifiers, but it can also detect other inputs, such as the templates used for comparative modeling. Thus, it is important for deposition that all input files are annotated as well as possible:
HEADER and TITLE lines.REMARK records and make sure that any files mentioned in REMARK 6 ALIGNMENT: or REMARK 6 SCRIPT: records are available (modify the paths if necessary, for example if you moved the PDB file into a different directory from the modeling script and alignment).EXPDTA and TITLE records to the files for ProtocolOutput to pick up. See the python-ihm docs for more information.# data_fn: header in the GMM file points to it.ProtocolOutput attempts to automatically generate as much as possible of the mmCIF file, but there are some areas where manual intervention is necessary because the data is missing, or its guess was incorrect. This data can be corrected by manipulating the ihm.System object directly, at the end of the script. We will look at a few examples in this section.
For cross-linking experiments, the mmCIF file contains a description of the cross-linking reagent used. This information is not in the CSV file. It can be provided in the Python script with the linker argument to passed to the PMI CrossLinkingMassSpectrometryRestraint, but if this argument was missing (or incorrect) we can correct it here. By checking the publications for the cross-link datasets from Al Burlingame's lab and Juri Rappsilber's lab), we can determine that the DSS and BS3 cross-linkers were used, respectively. These are common enough cross-linkers that python-ihm already includes definitions for them in the ihm.cross_linkers module (for less common linkers we can create an ihm.ChemDescriptor object from scratch to describe its chemistry). Then we just set the linker type for each cross-linking restraint in the list of all restraints:
ProtocolOutput captures information on the actual sampling, storing it in ihm.protocol.Step objects. Here it correctly notes that we ran Monte Carlo to generate 20000 frames:
However, in many modeling scenarios the modeling script is run multiple times on a compute cluster to generate several independent trajectories which are then combined. ProtocolOutput cannot know whether this happened. It is straightforward though to use the python-ihm API to manually change the number of output models to match that reported in the publication:
The current mmCIF file contains all of the input data, and notes that Monte Carlo was used to generate frames, but doesn't actually store any of those coordinates in the file. Ultimately this information will be added automatically by PMI model analysis and validation scripts, but for the time being any information about clustering, localization densities, and final models needs to be added to the file using the python-ihm API:
python-ihm allows for models to be placed into groups (ihm.model.ModelGroup) so we can make such a group for the cluster. These groups are in turn grouped in an ihm.model.State, a distinct state of the system (e.g. open/closed form, or reactant/product in a chemical reaction). Finally, states can themselves be grouped into ihm.model.StateGroup objects. All of these groups act like regular Python lists. In this case we have only one state, so we just add our model group to that:
Next, we can describe the cluster using the ihm.model.Ensemble class, noting that it was generated from our clustering:
To add a model to the file, we create an ihm.model.Model object, add atoms or coarse-grained objects to it, and then add that to the previously-created ModelGroup. ProtocolOutput provides a convenience function add_model which does this, converting the current IMP structure to python-ihm (we just need to load an IMP structure first, e.g. from an RMF file using IMP.rmf.load_frame).
In practice we typically want to store more than just a single model in the file, since a single model cannot represent the flexibility and variability of the complete ensemble. mmCIF files allow for storing multiple conformations (like a traditional PDB file for storing an NMR ensemble) and also representations of the entire ensemble (localization densities).
However, mmCIF is a text format and is not best suited for storing large numbers of structures. We can instead store the coordinates in a simpler, binary file and link to it from the mmCIF using the ihm.location.OutputFileLocation class, providing one or more representative structures (such as the cluster centroid) in the mmCIF file itself. DCD is one such format that we can (ab)use for this purpose (it is really designed for atomic trajectories). python-ihm provides an ihm.model.DCDWriter class which, given an ihm.model.Model, will output DCD.
If we have localization densities from the analysis for the cluster, we can add those to the Ensemble as ihm.model.LocalizationDensity objects. These take python-ihm subunits (or subsets of them) as ihm.AsymUnit objects. ProtocolOutput provides an asym_units member, which is a Python dict, to get python-ihm subunits given PMI component names (such as "Rpb4.0"). For example, we can easily add the localization density of the Rpb4 subunit for the entire cluster to the file:
(We can also add information about the fit of the model to each of the restraints. Most python-ihm restraint objects contain a fits member which is a Python dict with ihm.model.Model objects as keys and some sort of restraint-fit object as values. For example, we could note that the cluster centroid model m was fit against the EM map by adding an ihm.restraint.EM3DRestraintFit object with the cross correlation coefficient, if known. We could also specify the Bayesian nuisances ψ and σ for our cross-links with ihm.restraint.CrossLinkFit objects.)
We added paths for localization densities and the DCD file with ihm.location.FileLocation objects. These files won't be accessible to end users since they live on the local disk. (Recall from earlier that ProtocolOutput also adds local paths for the cross-link CSV files.)
FileLocation takes an optional repo argument which, if given, is an ihm.location.Repository describing the DOI where the files can be found, generally as a zip file (the first argument to ihm.location.FileLocation is a path within the repository if repo is specified, otherwise a path on the local disk). So we could explicitly refer to a DOI for each of our external files. (A number of services provide DOIs for uploaded files, such as Zenodo or FigShare.)
An alternative is to retroactively change the existing FileLocations (both those created automatically by ProtocolOutput and manually by us using python-ihm) using the root and top_directory arguments to Repository and the ihm.System.update_locations_in_repositories function:
This assumes that the zipfile at the given DOI and URL was created by archiving all files under the top directory of the GitHub repository (../.. relative to the directory containing the script). (It was actually created automatically by Zenodo when a new release of this tutorial was created on GitHub. Zenodo puts all the files in the zipfile under a uniquely-named directory, so we need to set top_directory to match.)
In principle archiving a GitHub repository at Zenodo is as simple as downloading the repository from GitHub as a zip file, then uploading that file to Zenodo. In practice, however, these files might be quite large, and so viewers such as ChimeraX may need to download a multi-gigabyte zip file just to get a relatively small input, such as a CSV or FASTA file. To remedy this, the repository can be split into multiple smaller zip files. See for example the multiple files archiving the 2018 NPC study; these were generated using this script.
Thus the cross-link file which was previously linked with the local path ../data/polii_xlinks.csv can now be found by downloading imp_deposition_tutorial-v0.2.zip and extracting salilab-imp_deposition_tutorial-1ad5919/rnapolii/data/polii_xlinks.csv from it:
We can now save the entire protocol to an mmCIF file (or to BinaryCIF, if we have the Python msgpack package installed) using the ihm.dumper.write function:
(BinaryCIF stores all the same data as mmCIF, but rather than storing it as a text file, the data is compressed and then written out as a MessagePack file, which is a form of binary JSON.)
mmCIF files can be viewed in many viewers. However, most viewers do not yet support the integrative modeling extensions, and so may only show the atomic parts of the model (if any). Integrative models can be viewed in ChimeraX - be sure to use a recent nightly build, and open the file from the ChimeraX command line using the format ihm option, e.g. open rnapolii.cif format ihm. (If you also want to see any DCD files, add ensembles true to the end of your command line to load them and then use the ChimeraX coordset command to manipulate the set of coordinates, e.g. coordset slider #1.3.2.)
ChimeraX also supports downloading and displaying structures directly from the PDB-Dev database, for example from the ChimeraX command line open 10 from pdbdev.
Note that even though PDB-Dev is still quite small, the models vary widely in composition, e.g.
- Some models (e.g. PDBDEV_00000018) are atomic and look much like a traditional "PDB" x-ray structure.
- Some models (e.g. PDBDEV_00000002) contain multiple states that have different sequences (this particular case contains exosome in nucleus-localized and cytoplasm-localized forms).
- Some models are not of proteins at all - e.g. PDBDEV_00000008 is a model of chromatin, part of the fly genome.
- Some models contain multiple representations - e.g. PDBDEV_00000012, a model of the yeast nuclear pore complex, contains both models of the scaffold (ring) and the flexible FG repeat regions which occupy the center of the pore.
VMD is also reportedly working on support in their forthcoming 1.9.4 release.
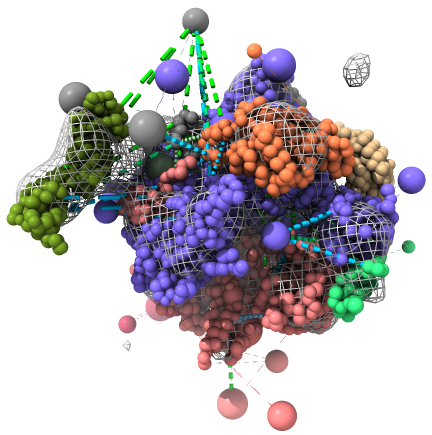
An example view of the deposition, as rendered by ChimeraX, is shown below. The EM map is shown as a mesh, the DSS cross-links as green dashed lines, and the BS3 cross-links as blue dashed lines. ChimeraX also shows the comparative models used as starting guesses for the subunit structures - toggle off the display of the result models to see them.
mmCIF files can also be viewed in a web browser, using Mol* Viewer. (Note that it currently only shows the coordinates, not the experimental information or other metadata.)
mmCIF files are also just plain text files, and can be viewed in any text editor (for example to check for errors, particularly for categories that ChimeraX doesn't support yet). Most of the data is stored as simple tables (look for the loop_ keyword in the file). For example, the coordinates of the coarse-grained beads are stored in the _ihm_sphere_obj_site table, the start of which in rnapolii.cif looks like:
This simply states that a sphere representing residue 1 in chain A is centered at (158.180, 180.876, 96.861) and has radius 3.068, and so on.
We can also read in mmCIF or BinaryCIF files using python-ihm's ihm.reader module and explore the data using the python-ihm API. For example, we can read the structure we just created:
Similarly we could explore any integrative model deposited in PDB-Dev. For example we can look at PDB-Dev #14, a HADDOCK model:
More tutorials on using IMP are available at the IMP web site.