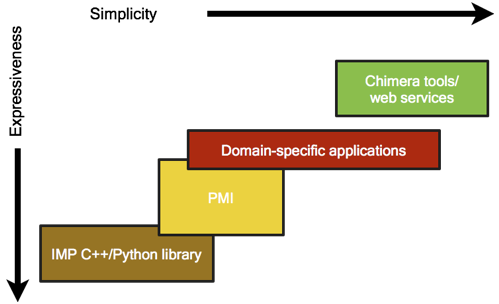
The Integrative Modeling Platform (IMP) software implements the integrative modeling procedure described above. Integrative modeling problems vary in size and scope, and thus IMP offers a great deal of flexibility and several abstraction levels as part of a multi-tiered platform:
The next parts of the manual will cover the use of the various parts of IMP, starting from the simplest:
- Chimera tools/web services: we provide a number of web services that use IMP at https://salilab.org/. Additionally, the UCSF Chimera software includes several tools that use IMP. These are the simplest to use because they do not require an IMP installation.
- Domain-specific applications: we provide a number of command-line tools, designed to be used by IMP users with no programming experience, that provide user-friendly applications to handle specific tasks, such as fitting of proteins into a density map of their assembly, or comparing a structure with the corresponding SAXS profile.
- PMI: the Python Modeling Interface (PMI) is a powerful set of tools designed to handle all steps of the modeling protocol for typical modeling problems. It is designed to be used by writing a set of Python scripts.
- IMP C++/Python library: at the lowest level, IMP provides building blocks and tools to allow methods developers to convert data from new experimental methods into spatial restraints, to implement optimization and analysis techniques, and to implement an integrative modeling procedure from scratch; the developer can use the C++ and Python programming languages to achieve these tasks.