 |
|
IMP Manual
for IMP version 2.6.1
|
The Python Modeling Interface (PMI) is a powerful set of tools designed to handle all steps of the modeling protocol for typical modeling problems. It is designed to be used by writing a set of Python scripts.
We will illustrate the use of PMI by determining the localization of two subunits of RNA Polymerase II, utilizing chemical cross-linking coupled with mass spectrometry, negative-stain electron microscopy (EM), and x-ray crystallography data. We will try to reconstruct the stalk of the complex, comprising of subunits Rpb4 and Rpb7, hypothesizing that we know already the structure of the remaining 10-subunit complex. The example can be easily generalized to any other set of subunits.
To work through the example on your own system, you will need the following packages installed in addition to IMP itself:
(If you are using Anaconda Python, you can get the Python packages above by simply running conda install numpy scipy scikit-learn matplotlib. On a Mac you can get them using the pip tool, e.g. by running a command like sudo easy_install pip, then install the packages with something like sudo pip install scikit-learn; sudo pip install matplotlib. numpy and scipy are already installed on modern Macs. Something similar may also work on a Linux box, although it's probably better to install the packages using the distribution's package manager, such as yum or apt-get.)
Then download the input files, either by cloning the GitHub repository or by downloading the zip file.
The rnapolii example contains three directories: analysis, data and modeling.
RNA Pol II is a eukaryotic complex that catalyzes DNA transcription to synthesize mRNA strands. Eukaryotic RNA polymerase II contains 12 subunits, Rpb1 to Rpb12. The yeast RNA Pol II dissociates into a 10-subunit core and a Rpb4/Rpb7 heterodimer. Rpb4 and Rpb7 are conserved from yeast to humans, and form a stalk-like protrusion extending from the main body of the RNA Pol II complex.
This example will use data from chemical cross linking, EM and x-ray crystallography to localize the two subunits of the RNA Polymerase II stalk (Rpb4, Rpb7) to a static core of the remaining ten subunits.
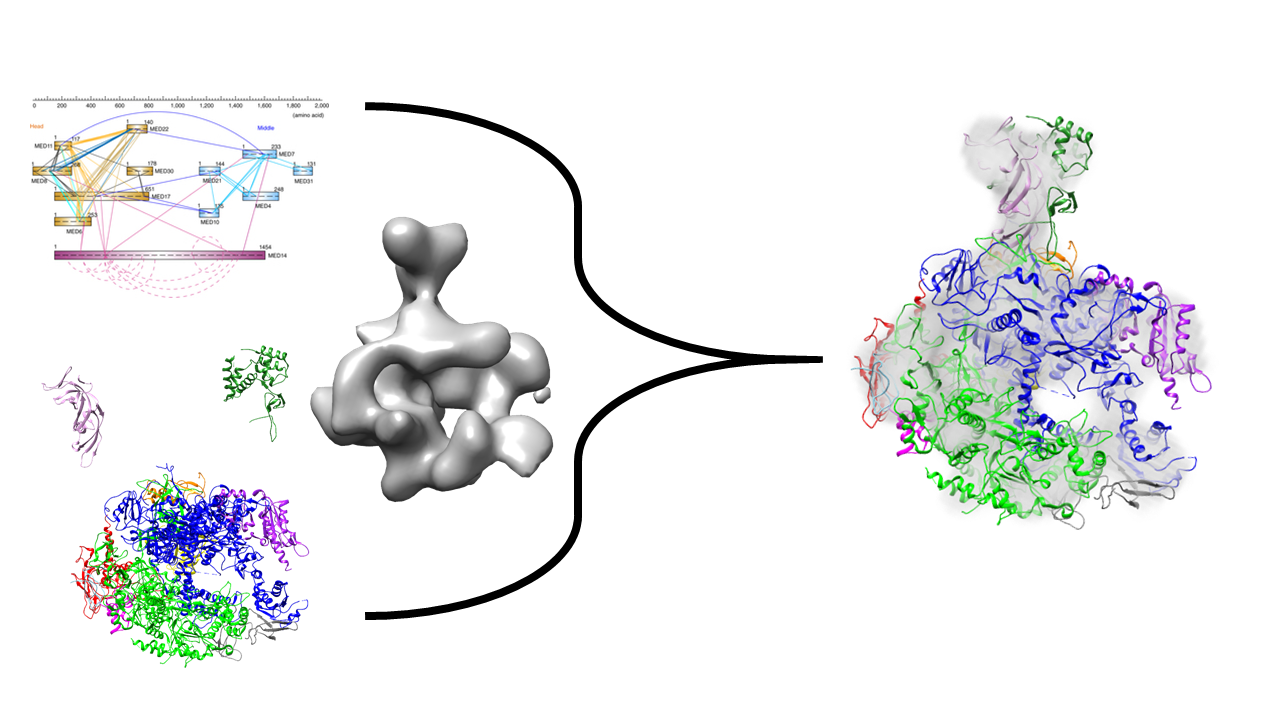
Structural modeling using IMP is divided into four stages.
Click the links below to see a breakdown of all the modeling steps.
The first three modeling stages are all contained within one script, modeling.py. You can get started by simply changing into the rnapolii/modeling directory and then running the script with Python:
It will take a very long time to complete the sampling; to get an idea of what's going on you can run it with only 50 output frames by adding the --test option: