Integrative structure determination of the yeast NPC
Summary
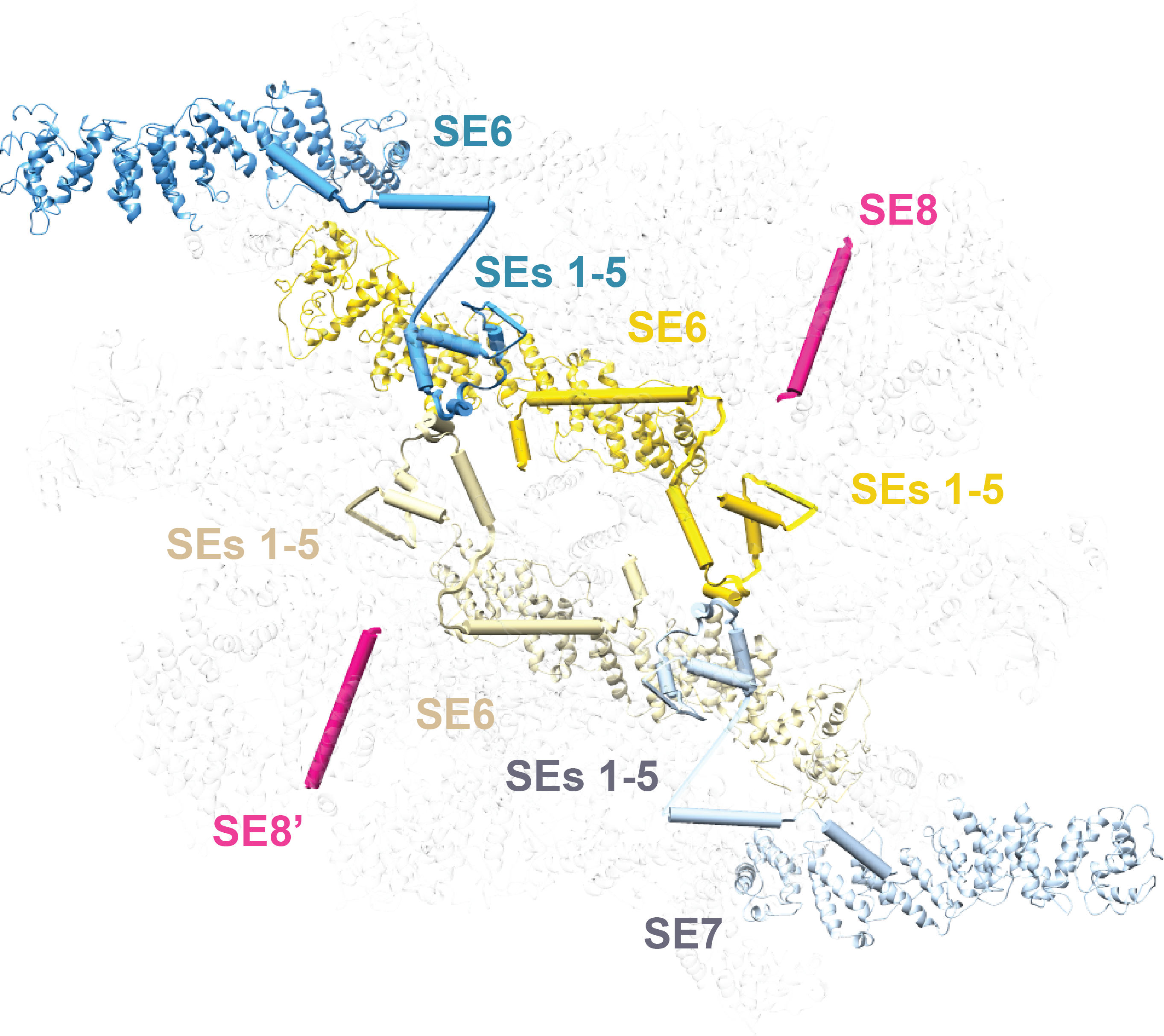
Nuclear pore complexes (NPCs) mediate the nucleocytoplasmic transport of macromolecules. Here we provide a structure of the isolated yeast NPC in which the inner ring is resolved by cryo-EM at sub-nanometer resolution to show how flexible connectors tie together different structural and functional layers. However, at the current resolution, orphan densities were revealed in the spoke that could not be accounted for by docked models. The density of these orphans is similar to neighboring Nups which indicates a high occupancy. We identified three orphans (group of 3) that interact with 3-helix bundles in the diagonal walls, and a gap at the center of the spoke is filled by 4 orphans arranged in two groups of 2. We also identified 3 orphan helices that bind to Nup192 and two orphan helices that bind to Nup188, which are present in two copies per spoke. In total, 30 orphan densities were mapped in the spoke that arise from flexible connectors. To solve the identity and connectivity of these orphans, we applied an integrative threading approach (Saltzberg et al., 2019) to compute Nup identity, copy identifier, and sequence segments for structure elements (SEs) in each orphan. This resulted in 28 orphan SEs being confidently assigned to contiguous sequences in four Nic96 N-terminal domains (NTDs).
Additionally, we used integrative modeling to determine the structure of the lumenal Pom152 ring in its restricted and dilated form.
List of files and directories
datacontains all relevant information: experimental data, statistical analyses, and prior information.integrative_structure_modelingcontains all the modeling script and results of the lumenal pom152 ring in the restricted and dilated forms.integrative_threadingcontains all modeling scripts of resultsSI_tablescripts to summarize the integrative modeling protocol into a single tableutilsother scripts necessary for data sharing
Information
Author (s): Ignacia Echeverria
License: CC BY-SA 4.0 This work is licensed under the Creative Commons Attribution-ShareAlike 4.0 International License.
Publications:
Akey CW, Singh D, Ouch C, Echeverria I, Nudelman I, Varberg JM, Yu Z, Fang F, Shi Y, Wang J, Salzberg D, Song K, Xu C, Gumbart JC, Suslov S, Unruh J, Jaspersen SL, Chait BT, Sali A, Fernandez-Martinez J, Ludtke SJ, Villa E, Rout MP. Comprehensive structure and functional adaptations of the yeast nuclear pore complex. Cell. 2022 Jan 20;185(2):361-378.e25. doi: 10.1016/j.cell.2021.12.015. Epub 2022 Jan 3. PMID: 34982960; PMCID: PMC8928745. [LINK] (https://www.sciencedirect.com/science/article/pii/S0092867421014537?via%3Dihub)


 Download files
Download files Warning: these files have not yet been verified to work with the latest version of IMP. We will update this page when they have been.
The files are also available at
Warning: these files have not yet been verified to work with the latest version of IMP. We will update this page when they have been.
The files are also available at  To install the software needed to reproduce this system with the
To install the software needed to reproduce this system with the
 To set up the environment on the UCSF Wynton cluster to run
this system, run:
To set up the environment on the UCSF Wynton cluster to run
this system, run: