 |
IMP Tutorial
for IMP version 2.0.1
|
 |
IMP Tutorial
for IMP version 2.0.1
|
To understand the function of a macromolecular assembly, we must know the structure of its components and the interactions between them. However, direct experimental determination of such a structure is generally rather difficult. While multiple methods do exist for structure determination, each has a drawback. For example, crystals suitable for X-ray crystallography cannot always be produced, especially for large assemblies of multiple components. Cryo-electron microscopy (cryo-EM), on the other hand, can be used to study large assemblies, but it is generally limited to worse than atomic resolution. Finally, proteomics techniques, such as yeast two-hybrid and mass spectrometry, yield information about the interactions between proteins, but not the positions of these proteins within the assembly or the structures of the proteins themselves.
One approach to solve the structures of proteins and their assemblies is by integrative modeling, in which information from different methods is considered simultaneously during the modeling procedure. The approach is briefly outlined here for clarity; it has been covered in greater detail previously. These methods can include experimental techniques, such as X-ray crystallography, nuclear magnetic resonance (NMR) spectroscopy, electron microscopy (EM), footprinting, chemical cross-linking, FRET spectroscopy, small angle X-ray scattering (SAXS), and proteomics. Theoretical sources of information about the assembly can also be incorporated, such as template structures used in comparative modeling, scoring functions used in molecular docking, as well as other statistical preferences and physics-based energy functions. Different methods yield information about different aspects of structure and at different levels of resolution. For example, atomic resolution structures may be available for individual proteins in the assembly; in other cases, only their approximate size, approximate shape, or interactions with other proteins may be known. Thus, integrative modeling techniques generate models at the resolution that is consistent with the input information. An example of a simple integrative approach is building a pseudo-atomic model of a large assembly, such as the 26S proteasome, by fitting atomic structures of its subunits predicted by comparative protein structure modeling into a density map determined by cryo-EM.
The integrative modeling procedure used here is shown below.
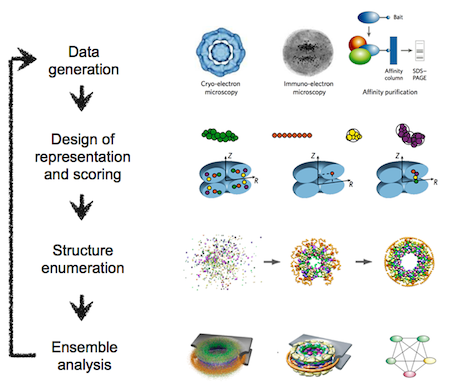
The first step in the procedure is to collect all experimental, statistical, and physical information that describes the system of interest. A suitable representation for the system is then chosen and the available information is translated to a set of spatial restraints on the components of the system. For example, in the case of characterizing the molecular architecture of the nuclear pore complex (NPC), atomic structures of the protein subunits were not available, but the approximate size and shape of each protein was known, so each protein was represented as a ‘string’ of connected spheres consistent with the protein size and shape. A simple distance between two proteins can be restrained by a harmonic function of the distance, while the fit of a model into a 3D cryo-EM density map can be restrained by the cross-correlation between the map and the computed density of the model. Next, the spatial restraints are summed into a single scoring function that can be sampled using a variety of optimizers, such as conjugate gradients, molecular dynamics, Monte Carlo, and inference-based methods. This sampling generates an ensemble of models that are as consistent with the input information as possible. In the final step, the ensemble is analyzed to determine, for example, whether all of the restraints have been satisfied or certain subsets of data conflict with others. The analysis may generate a consensus model, such as the probability density for the location of each subunit in the assembly.
The Integrative Modeling Platform (IMP) software implements the integrative modeling procedure described above. Integrative modeling problems vary in size and scope, and thus IMP offers a great deal of flexibility and several abstraction levels as part of a multi-tiered platform:
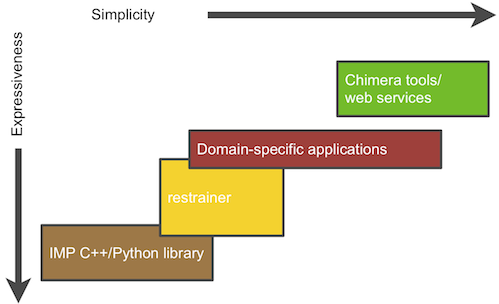
At the lowest level, IMP provides building blocks and tools to allow methods developers to convert data from new experimental methods into spatial restraints, to implement optimization and analysis techniques, and to implement an integrative modeling procedure from scratch; the developer can use the C++ and Python programming languages to achieve these tasks. Higher abstraction levels, designed to be used by IMP users with no programming experience, provide less flexible but more user-friendly applications to handle specific tasks, such as fitting of proteins into a density map of their assembly, or comparing a structure with the corresponding SAXS profile.
In the next part of the tutorial, we will demonstrate the building blocks and tools in the lowest level of the IMP software, then move to the higher level tools in later parts.